Start a new Ion AmpliSeq™ DNA Gene design
DNA Gene designs can include a mix of gene and region targets, including small hotspot regions. If you are using one of the Ion AmpliSeq™ standard genome references, you can design a custom DNA gene panel to discover new variants as well as detect known SNPs and INDELs in your genes of interest.
- In the home screen, in the Ion AmpliSeq™ Custom panels pane, click Create Custom panel.
- In the Start a new Ion AmpliSeq™ design screen, in the Name and details step, enter a design name and optional details.
-
In the
Application Type
step, click
DNA Gene designs (multi‑pool).
If you are interested in CNV detection, see Ion AmpliSeq panel design and CNV detection.
Selecting the application type filters the genomes to display only the selections that are compatible.
-
In the
DNA Type step,
select a DNA type that is appropriate for your sequencing experiment.
Note: This selection affects amplicon length in the design.
- In the Select genome to use step, select a reference.
-
Click Next: Add Targets.
A draft design screen opens (see Figure 1).
-
Add targets using the options available in
the target addition pane (see callout 1 in Figure 1).
For guidelines on how to add targets, see Ion AmpliSeq Custom panels: DNA gene design input specifications.
Option
Description
Add targets manually.
-
In the Add Gene/Region tab, in the Type column, select Gene (CDS only), Gene (CDS +UTR), or Region.
-
In the Gene Symbol field, enter a gene symbol. Alternatively, start typing the gene symbol or region, then select the gene from the dropdown list.
-
Click Add target after each entry.
If a target was added successfully, the Target saved successfully message appears. If a target name or symbol is invalid, the Target could not be saved error message appears. If needed, you can correct erroneous targets in the targets table, as described in step 8.
In the Add Amplicon by ID field, enter the amplicon IDs assigned to specific genomic coordinates, then click Add amplicon.
Upload genomic coordinates of multiple targets at one time using a CSV or BED file.
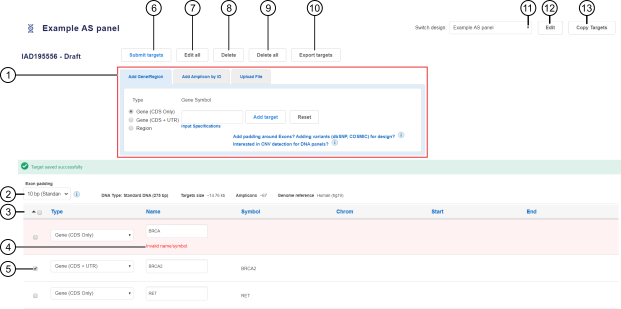 Figure 1. Draft design
screen
Figure 1. Draft design
screen
-
A target addition pane with multiple methods for adding targets to a design. Each tab contains fields that are specific to the corresponding target addition method.
-
Customize exon padding.
-
Targets table lists all added targets in a design.
-
Targets that did not pass verification are marked in red in the targets table. Fields that contain errors are marked with Invalid and can be corrected.
-
Select the checkbox in the row of each target that you want to delete or edit.
-
Click to submit a design.
-
Click to change gene type for selected targets (see callout 5) or all targets.
-
Click to delete the selected targets (see callout 5).
-
Click to delete all targets.
-
Click to export all targets to a CSV file.
-
Select from the dropdown list to display a different design.
-
Click to edit design name and details.
-
Click to copy targets to a new custom design.
The software uploads the targets, checks them, and verifies regions. The targets are added to the targets table (see callout 3 in Figure 1).
-
-
Review and modify the design as
needed.
Action
Description
Correct erroneous targets
Targets that contain errors are highlighted in red in the targets table. In the row of each erroneous target, modify the fields that are marked as Invalid (see callout 4 in Figure 1). Invalid fields can include Name/Symbol, Chrom, Start, and End.
Delete targets
In the targets table, select the checkbox in the row of one or more targets, then click Delete. To delete all targets, click Delete all. See callouts 5, 8, and 9 in Figure 1.
Customize exon padding
The standard exon padding is 10 bp. If you need to provide a custom padding between 0 and 100 bases around the exons in your design, select the desired value from the Exon padding dropdown list (see callout 2 in Figure 1).
IMPORTANT! The selected value applies to both before and after the exon, and is applied across all exons in a design.
Change gene type
You can modify gene type for each individual target in the targets table, or use the Edit all functionality to edit multiple targets at one time.
-
To change the gene type for one target, in the targets table, in the row of a target, in the Type column, select a gene type from the dropdown list (see callout 3 in Figure 1).
-
To change the gene type for multiple targets at once, in the targets table, select the targets that you want to modify, then click Edit all. In the Edit All dialog, select the gene type from the dropdown list, then click Apply to Selected or Apply to All. See callouts 5 and 7 in Figure 1.
Export targets to a CSV file
Click Export targets. A CSV file that contains the list of all added targets is automatically downloaded to your local storage (see callout 10 in Figure 1).
Edit design name and details
Click Edit, then in the Edit your design screen, modify design name and details as needed. Click Save. See callout 12 in Figure 1.
Copy targets to a new design
Click Copy Targets, then in the Copy Design dialog, select the panel type and DNA type, and enter the name and details for the new design. Click Save. See callout 13 in Figure 1.
The new design is added to the list of your custom designs. To view or edit the new design, click in the navigation bar. Alternatively, open a new design by selecting a design from the Switch design dropdown list (see callout 11 in Figure 1). For designs that have been submitted, or locked, the design opens to the Results ready screen.
-
-
To submit your panel, click
Submit targets,
then click
OK to
confirm your submission.
Up to 5 designs can be submitted at one time.
After submission, a submitted design screen opens, displaying the design confirmation message.
After you submit your design, the targets are locked ( ) and can no longer be edited.
When the assay design results are ready, you receive an e‑mail notification
instructing you to review the results in Ion AmpliSeq™ Designer. Click the
View results
link provided in the email to be directed to the results page. Alternatively, sign
in to Ion AmpliSeq™ Designer (AmpliSeq.com), then in the navigation bar
click to access the results for your design (see My Designs).
) and can no longer be edited.
When the assay design results are ready, you receive an e‑mail notification
instructing you to review the results in Ion AmpliSeq™ Designer. Click the
View results
link provided in the email to be directed to the results page. Alternatively, sign
in to Ion AmpliSeq™ Designer (AmpliSeq.com), then in the navigation bar
click to access the results for your design (see My Designs).
To review design details and place an order, proceed to Order an Ion AmpliSeq custom panel.
