The Hotspots Pipeline
Single pool designs represent cost and handling advantages over panels that require two or more pools of primers. Designs targeting small genomic regions such as SNPs and small indels are well suited to fit in one single pool since those regions can be covered by non-overlapping amplicons.
In regular designs the “tiling” stage of the AmpliSeq pipeline is in charge of selecting amplicons to cover the target regions. Since very often the regions are larger than a single amplicon, several amplicons are required to cover one particular target. This process results in a set of overlapping amplicons that, if put together, are prone to chemically interact with each other during the sequencing process. The “pooling” stage is in charge of creating as many pools as needed to avoid these interactions.
The hotspots pipeline includes special algorithms that in the pooling step allow the creation of one single pool of primers with maximum coverage for targets as small as one single base and as large as 50.
The idea behind the hotspots designer can be better understood by looking at the following diagram in which the long line represents a genomic region; a, b and c are the amplicons available for covering that region. The red portions of the amplicons represent the primers.
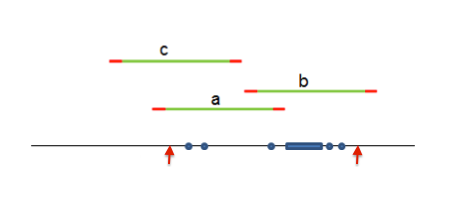
In a regular (non-hotspots) AmpliSeq panel, the targets are long regions (delimited by arrows in the diagram). The algorithm attempts to find a set of primers of minimum cost to cover the target (amplicons a and b). Amplicons a and b overlap so this would require 2 pools to cover the region in question. Forcing the panel to be in one pool, would either eliminate amplicon a or b resulting in reduced coverage for the 1 pool solution. Using amplicons c and b is not an option since they fail to cover a portion of the region between the arrows. In contrast, for a hotspots panel the targets are small genomic regions (marked with dots over the line representing the genome), the hotspots pipeline would actually select non-overlapping amplicons c and b to cover the submitted targets. Non-overlapping amplicons can be all put together in a single pool of primers.








