Release Notes
7.8.12 (17 September 2025)
Application area field for custom panels
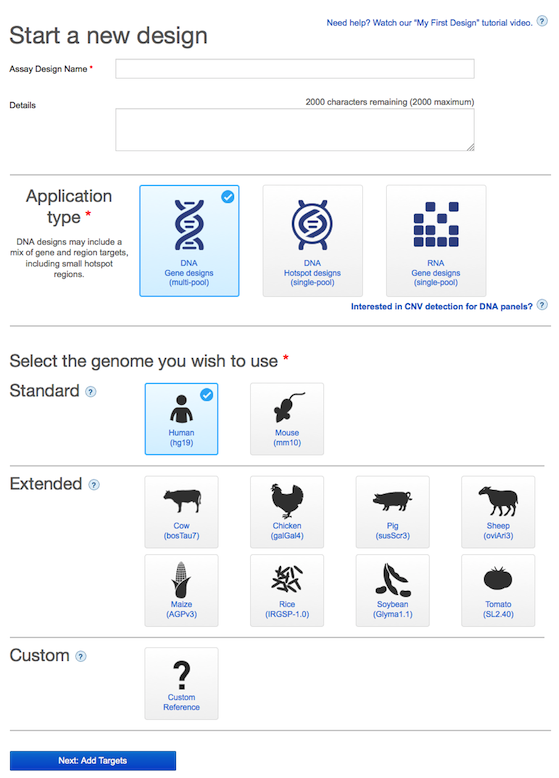
In this release, we have introduced an application area design attribute that allows users to specify the intended application area when creating their custom Ion AmpliSeq™ Made-To-Order, Oncomine™ tumor specific, Ion AmpliSeq On-Demand, and Ion AmpliSeq HD designs. A dropdown menu is now available in the first step of creating new custom designs, where users are required to select one of the available application area options or enter their own custom value. The selection made during design creation will remain associated with each design and can be used to facilitate navigation of the available custom designs by application area in the My Designs dashboard, where a filter has also been implemented for this purpose.
The options available in the application area dropdown menu are as follows:
- Cancer Research (Solid Tumor/Tissue Sample)
- Cancer Research (Liquid Biopsy Samples)
- Cancer Research (Heme)
- Cancer Research (Inherited Cancers)
- Reproductive Health Research
- Genetic Disorders Research
- Microbial/Infectious Disease Research
- Pharmacogenomics Research
- Human Identification Research
- Agricultural/Breeding Research
- Other Research (free text)
Notes: For custom designs created before this release, the application area will be left unselected. Users can optionally set a value from a dropdown menu in the Edit Design functionality available on each design page. Once set, the application area value for a given panel cannot be modified.
We have added information icons for each custom product line on the Home Page. When clicked, these icons display short on-page tooltips that provide brief descriptions of the main characteristics of each custom panel type and links to more detailed informative pages. This feature is designed to help users select the right custom panel type for their research purposes.
Additionally, we have introduced a "Contact Sales" button on the website's top bar. This button allows users to submit commercial queries related to Ion AmpliSeq panels through a web form. Based on the information provided in the form, requests will be redirected to the appropriate commercial representative, who will follow up with the request.
Product launches: Expansion Panels
We introduced Ion AmpliSeq Expansion Panels, a custom panel category for internally developed and verified custom Ion AmpliSeq spike-in panels meant to expand the content of available Oncomine and other Ion AmpliSeq assays.
On July 1st we launched the following two Ion AmpliSeq Expansion Panels for the Oncomine Myeloid DNA Assays:
Navigate to the corresponding design page or contact Support to learn more.
Known limitations:
- The Help documentation is currently under revision for the latest features and is expected to be updated in future AmpliSeq Designer releases.
- UCSC is not displaying tracks for all custom Made-to-Order designs.
7.8.11 (17 June 2025)
COSMIC v97 and dbSNP v156 database update
In this release we have updated our support for the COSMIC database to version v97 and dbSNP database to version v156 for both hg19 and hg38 human reference genomes. Targets entered as COSMIC or dbSNP identifiers in hotspot designs (Made-To-Order and Made-To-Order HD) will refer to these versions of the respective database.
Notes: Support for older COSMIC and dbSNP versions is not available. Users can enter genomic coordinates of obsolete COSMIC or dbSNP annotations if needed.
Known limitations:
The Help documentation is currently under revision for the latest features and will be updated in future AmpliSeq Designer releases.
7.8.8 (15 April 2025)
This release adds a new informational field to the Order Preview page for Made-To-Order, White Glove, and Community panels. This field indicates the number of samples that can be processed per order of the panel based on the selected instrument, library prep, format, and special instructions configuration. The provided numbers assume no dead volume and represent a maximum theoretical value.
Notes: The samples per order information is not currently implemented for AmpliSeq HD panels and small pack formats for selected Community panels.
Improved Clarity on Provided Materials Information in Order Preview Page
We have enhanced the "Provided Materials" field in the Order Preview page for Made-To-Order, AmpliSeq HD, Community, and White Glove panels. This update clarifies the actual fill volumes delivered for each panel configuration, ensuring that users have precise and detailed information about the materials included with their order.
Minor bug fix
We have addressed a minor bug that, in rare instances, prevented users from proceeding with the panel Order Preview on ampliseq.com due to incomplete information in their thermofisher.com account data. With this fix, users with incomplete account information can still configure for order, add to cart and request quotes for their panels.
Please note that while this fix at the ampliseq.com level allows users to perform these actions, completing thermofisher.com account data is still required to proceed to order check out and/or receive web quotes.
Known limitations:
The Help documentation is currently under revision for the latest features and will be updated in future AmpliSeq Designer releases.
7.8.7 (January 28, 2025)
Send SOS Document by email from Order Preview Page
The Standard Order Sheet (SOS) document captures panel order configuration details in a PDF file from the Order Preview page and can be sent offline as needed (e.g., to Sales Representatives or Customer Service). This release introduces the ability to email the SOS document directly from the Order Preview page.
When clicking the new "Generate SOS" button on the Order Preview page, users can now choose to send the SOS document by email in addition to downloading it. Upon selecting this option, a pop-up appears, allowing users to enter the recipient's email address(es) and an optional personalized message*. The email, with the SOS document attached, is sent from ionampliseq-noreply@lifetech.com.
Notes:
- A copy of the SOS document email is also sent to the user's login email address for confirmation.
- Recipients may need to check their spam folders and whitelist the sender address if the email is not delivered within a few minutes.
*The SOS document email is auto-generated from a no-reply address. Consider including specific contact details in the personalized message field as necessary.
Known limitations:
- The Help documentation is currently under revision for the latest features and will be updated in future AmpliSeq Designer releases.
7.8.6 (December 03, 2024)
Enhanced Copy Amplicon Functionality
The copy amplicon feature on AmpliSeq.com allows users to combine amplicons from existing panels (source designs) into new full (only copied amplicons) or partial (additional content added) subset panels. For source designs with multiple pools, amplicons are automatically reassigned to pools, resulting in different pool distributions.
This release introduces a new pool preservation option that ensures copied amplicons retain their original pool assignments in subset designs. This option is available for compatible multi-pool designs* through the direct amplicon copy process (design-to-design) and is limited to one source design. Both full and partial subset designs support pool preservation.
Amplicons are selected by target from the source design page and single copied amplicons can optionally be removed while the subset design is in draft stage (before submission). This might potentially lead to subset designs with imbalanced pools and users should always verify pool balancing in the final subset design and contact Technical Support if needed.
In addition to pool preservation, this release also introduces an enhanced user interface behavior when combining amplicons from source designs with different amplicon sizes.
*Compatible designs include Made-To-Order, certain White Glove designs, human pre-designed panels, and custom AmpliSeq HD designs. Logic and exceptions may vary for specific design types.
Small pack size available for Ion AmpliSeq™ Liverpool Lymphoid Network Panel
Effective October 29, 2024, the Ion AmpliSeq™ Liverpool Lymphoid Network Panel is also available in a smaller pack size. This new format can be selected during the order preview and consists of 1.5 mL of pooled amplicons per pool, accommodating 80 samples on GeneStudio/Chef DL8 and 40 samples on Genexus under recommended run configurations (assumes no dead volume).
Known limitations:
- The Help documentation is currently under revision for the latest features and will be updated in future AmpliSeq Designer releases.
7.8.5 (September 12, 2024)
Enhanced design pipeline for AmpliSeq HD designs
A new design pipeline improvement was introduced for custom AmpliSeq HD designs with the scope of maximizing the utilization of shorter HD amplicons whenever available, without impacting the overall number of amplicons or the in silico coverage of the design. The improvement applies to both AmpliSeq HD amplicon size ranges (75-125bp for cfDNA/FFPE dual use, and 125-175bp for FFPE materials). The preference for shorter amplicons in AmpliSeq HD designs is intended to overall promote a better target amplification efficiency on low-quality / short size input materials.
Starting from this release, the SOS document capturing the order format information for all custom panel types for the intended purposes will be provided in pdf format instead of txt format, for better document readability, handling and compatibility. The content and scope of the SOS document remain otherwise unchanged.
Known limitations:
- The Help documentation is currently under revision for the latest features and will be updated in future AmpliSeq Designer releases.
7.8.4 (July 30, 2024)
Redesign of Pre-Cart page for Ion AmpliSeq Community and White Glove panels
We have applied the "Pre-Cart" page look and feel introduced in release 7.8.3 for AmpliSeq and AmpliSeq HD Made-To-Order panels also to Community and White Glove panels. This new interface has redesigned informational tables and the functionality to add compatible consumables to the panel order cart organized in dynamic accordions based on consumable categories.
Known limitations:
- The Help documentation is currently under revision for the latest features and will be updated in future AmpliSeq Designer releases.
7.8.3 (June 12, 2024)
Redesign of Made-To-Order panel pages
We have revised the user interface of the main functional pages related to AmpliSeq standard and AmpliSeq HD Made-To-Order panels to overall improve the user experience with panel design, customization, and ordering. Improved pages include:
- "Start new AmpliSeq" page:
- Reorganization of the start new panel design workflow in logical order.
- Better alignment of the new panel design workflow items.
- "Design draft" and "Results-ready" pages:
- Target and amplicon action functionalities accessible through dedicated buttons.
- Advanced column sorting and dynamic filtering capabilities in panel content tables.
- Intuitive graphic representation of design in-silico coverage.
- A new "More Actions" button grouping the following design functionalities: "Propose as Community Panel", "Edit Description", "Delete Design", "Amplicon Histogram Download", "Sharing", and "Chip Calculator".
- Design results displayed through improved dynamic accordions for hotspots, RNA fusions and RNA gene expression designs.
- "Pre-Cart" page:
- Redesigned informational tables.
- Functionality to add compatible consumables to the panel order cart organized in dynamic accordions based on consumable categories.
Automatic spike-in panel creation (compatible AmpliSeq DNA panels)
A new "Create Spike-in" button allows users to input targets for the automatic design of spike-in panels to expand the content of standard AmpliSeq DNA panels up to 2 pools. At present, the panels compatible with this feature include human DNA Community Panels and previously ordered AmpliSeq DNA Made-To-Order panels (any genome).
Once ready, automatic spike-in panels:
- Are accessible through a direct link from the page of the primary panel they were designed against (one spike-in per panel is allowed at any given time for custom Made-To-Order panels).
- Have amplicons automatically designed to be compatible with the existing amplicons in the primary panel (in terms of both pooling and amplicon length). A target bed file combining the primary and the spike-in panel content for analysis purposes is also automatically generated and made available in the results file of the spike-in panel.
- Can be either hotspot or gene designs type (while maintaining the same number of pools and the same reference genome of their primary panel for compatibility reasons).
- Can be ordered only as "50X spike-in" or "384-well plates only" formats for spike-in use purposes.
- Are automatically checked for the maximum number of amplicons per pool. Spike-in panels resulting in more than 123 amplicons per pool are marked as "incompatible" in the primary panel page and can still be ordered, but only in formats different from "50X spike-in" (to be used as standalone panels as needed).
Known limitations:
- The Help documentation is currently under revision for the latest features and will be updated in future AmpliSeq Designer releases.
7.8.2 (March 21, 2024)
Redesigned login page
The AmpliSeq.com login page was redesigned to communicate more clearly to users what AmpliSeq technology is, how it can be used, and the benefits it provides. The new page implements direct links to application, educational pages, and videos.
The access to Ion AmpliSeq Designer functionalities for panel customization remains otherwise unchanged (same login credentials).
✓ Ion AmpliSeq™ Liverpool Lymphoid Network Panel
The Ion AmpliSeq™ Liverpool Lymphoid Network Panel for hemato-oncology research is a targeted NGS community panel designed by leading clinical researchers at Liverpool Clinical Labs for the classification of lymphoid malignancy samples. This targeted panel covers 60 genes, providing comprehensive coverage of key biomarkers (including NOTCH1 3’UTR) applicable for the study of a range of lymphoid neoplasms, including both B and T cell lymphomas. This panel provides high accuracy NGS results with rapid turnaround time has one of the lowest DNA input requirements. Its performance has been verified by the network on both the GeneStudio™ and Genexus™ Integrated Sequencer Systems in a multicenter study using >200 real-world clinical research samples.
✓ Ion AmpliSeq™ Liverpool MPN Panel
The Ion AmpliSeq™ Liverpool MPN Panel for hemato-oncology research is a targeted NGS community panel designed by leading clinical researchers at Liverpool Clinical Labs for the classification of myeloproliferative neoplasm (MPN) samples. This panel provides high accuracy NGS results with rapid turnaround time, is compatible with the simple and automated Genexus workflow, and has one of the lowest DNA input requirements. A single pool design allows sequencing a high volume of samples (32 per lane per run) for cost-effective analysis. This DNA panel contains targets from the 3 classical MPN genes (JAK2, MPL, and CALR) and other key targets from 22 additional genes (including FLT3, IDH1, and IDH2) to provide a comprehensive assessment and classification of MPN samples in a single test. The panel’s performance has been verified using real-world clinical research samples and demonstrated 100% sensitivity across variant types, including challenging targets like FLT3-ITDs.
The panel is intended for Research Use Only and not for use in diagnostic procedures.
Known limitations:
- The Help documentation is currently under revision for the latest features and will be updated in future AmpliSeq Designer releases.
7.8.1 (January 17, 2024)
Starting from this release, the 50X Spike-in format is available also for the Genexus ordering option for compatible custom Made-To-Order DNA designs with less than 123 amplicons per pool. After selecting the Genexus instrument in the order preview process, users can now select the 50X spike-in format from a new special instruction dropdown menu. Spike-in panels are meant to be physically combined with existing primary panels to expand their content. Spike-in amplicons shall not overlap with existing primary panel amplicons to avoid unwanted interactions. At this time, users are responsible for such amplicon compatibility check (e.g., by visual inspection of panel target bed files). Additional support is available at Regional support contact links (Americas, EMEA, Greater China, South Asia, Japan).
Discontinuation of Ion PGM™, Ion OneTouch™2 and Ion Proton™ Systems
Kits for Ion PGM™, Ion OneTouch™ 2 (including GeneStudio™ S5 OneTouch™ 2 kits), and Ion Proton™ Systems have been discontinued on December 31st, 2023. Accordingly, in this release we have removed support for such kits and platforms from panel pages and documentations. Exceptions are all instances within the description text of Community Panel pages, where references were kept as integral part of the features of each panel at launch (actual instrument and chip compatibility information are available in the dedicated section of each page).
Known limitations:
- The Help documentation is currently under revision for the latest features and will be updated in future AmpliSeq Designer releases.
7.8 (November 7, 2023)
Automatic design of Made-To-Order panels compatible with gene-level CNV detection (human only)
Starting from this release, when creating human (hg19/hg38) Made-To-Order gene panels, gene targets (CDS and CDS+UTR) can be flagged for enabling automatic gene-level CNV detection compatible designs according to general AmpliSeq CNV design guidelines provided in the informational link available in the panel creation page. When this option is selected for a gene target, the design pipeline will try to ensure that at least 12 amplicons are automatically assigned to that target (if needed, amplicons will be added into the gene introns, if available, and then into upstream and downstream regions of the gene). Amplicons automatically added for CNV compatibility purposes will be listed in a specific bed file in the design results.zip file.
Once the design is completed, single CNV gene targets for which the minimum number of amplicons is reached are flagged as “CNV compatible” ( ), otherwise they are flagged as CNV incompatible (
), otherwise they are flagged as CNV incompatible ( ).
).
The overall design CNV compatibility status will also be checked against the general AmpliSeq CNV design guidelines and reported in the solution summary table (using the same symbols above). A design is considered CNV compatible if, and only if all the following conditions are met: 1) at least one of the requested CNV gene targets is CNV-compatible; 2) more than 60 amplicons in total exist in the panel; 3) the number of amplicons assigned to each CNV-compatible gene target is less than 40% of the total amplicons in the panel (as a proxy for the theoretical ideal condition at 20% per CNV target).
The specific reason(s) for design-level CNV incompatibility will be reported in a banner on panel page with the intent to help users identifying, when possible, alternative design strategies. Additional support is available at Regional contact links (Americas, EMEA, Greater China, South Asia, Japan).
Whereas Copy Number Variants can in principle be detected following design guidelines and best practices using compatible custom Ion AmpliSeq panels and the Ion Reporter Software, experimental testing, optimization, and verification are recommended. CNV design compatibility per se does not guarantee CNV detection performance.
New Ion AmpliSeq™ Newborn Screening Research Spike-In Panel for Severe Combined Immunodeficiency now available
In newborn screening (NBS) research, next-generation sequencing can provide genetic insights that aid in the understanding of severe combined immunodeficiency (SCID) and other T cell lymphopenias. Designed with leading researchers from the Department of Newborn Screening at Oslo University Hospital (Norway), the Ion AmpliSeq™ Newborn Screening Research Spike-In Panel for Severe Combined Immunodeficiency is a spike-in panel that, when combined with an AmpliSeq On-Demand primary panel created on ampliseq.com using the CSV file available from a direct link on the panel page, enables the investigation of 188 genetic targets for SCID, inborn errors of metabolism (IEM), congenital hypothyroidism (CH), congenital adrenal hyperplasia (CAH), cystic fibrosis (CF), T-cell defects, and relevant US NBS genes using dried blood spot (DBS) DNA samples.
This panel is customizable to meet your unique research needs directly on AmpliSeq.com. Contact regional AmpliSeq Support (Americas, EMEA, Greater China, South Asia, Japan) for further assistance.
The panel is intended for Research Use Only and not for use in diagnostic procedures.
Known limitations:
- The Help documentation is currently under revision for the latest features and will be updated in future AmpliSeq Designer releases.
7.6.10 (September 6, 2023)
Starting from this release standard AmpliSeq Made-To-Order designs with less than 12 amplicons per pool can be ordered to be used as spike-in panels. Accordingly, the only available special instruction ordering options for these panels will be “50X – spike-in” (Tubes of primers pools only format) and “384-well plates only” (Tubes plus 384 well plates format). Panels falling in this category will have banners in their panel page to inform users of the same.
Known limitations:
- The Help documentation will be updated in future AmpliSeq Designer releases for the latest features.
7.6.9 (July 18, 2023)
Support of new Genexus Software 6.8 features for custom assays
In this release we have updated AmpliSeq.com content to support new features introduced by Genexus Software 6.8 released on July 15th, 2023. These include:
Support for GX7 chip
Genexus Software 6.8 supports the new GX7 chip with custom AmpliSeq assays. GX7 chip enables 20-25M reads per lane (67% increase vs. GX5) expanding the sequencing capacity of the Genexus Integrated System.
Accordingly, we have updated Instrument(Chip) info in panel page, chip calculator, FAQ content and recommended consumables in Order Preview page with GX7 chip and supported consumables info.
Support for 48 Genexus AmpliSeq barcodes
The 48 Genexus AmpliSeq barcode plates introduced with Genexus Software 6.8 support automated AmpliSeq library preparation for custom panels for up to 48 libraries per run on both GX5 and GX7 chips, increasing the maximum library capacity per run by 50% and reducing sequencing costs per sample (in combination with the support for 6 reactions per strip 1-2 pair).
The new Genexus™ Barcodes 1-48 AS (Cat. A54129) and Genexus™ Barcodes 49-96 AS (Cat. A54130) are now listed for ordering among the recommended consumables in the Order Preview page.
Note: 48 Genexus AmpliSeq barcode plates do NOT support AmpliSeq HD library preparation.
Support for 24 samples per order for Ion AmpliSeq On-Demand panels for Genexus
When run on Genexus Software 6.8 (and above), all Ion AmpliSeq On-Demand panels for Genexus packages support library preparation for up to 24 samples per order (vs. previous 16 samples per order) when used in combination with 48 AmpliSeq Genexus barcode plates and when sequenced on a GX7 chip (regardless of the Genexus Barcode plate type used). The corresponding information has been updated for all Genexus AmpliSeq On-Demand formats in the Order Preview page.
Note: The price of AmpliSeq On-Demand panels for Genexus has remained unchanged, therefore reducing the panel price per sample by 33%.
Full support for Ion AmpliSeq HD 2-pool cfDNA assays on Genexus
Genexus Software 6.8 fully supports AmpliSeq HD 2-pool cfDNA panels both on GX5 and GX7 chips, allowing to investigate FFPE/liquid biopsy samples at full gene level with a single panel. Accordingly, we have now enabled the Genexus ordering option for AmpliSeq HD 2-pool cfDNA designs.
For more details on the use of custom AmpliSeq panels on Genexus 6.8, please refer to the new Ion AmpliSeq™ & Ion AmpliSeq™ HD Custom Assay User Guide (link to the same document is now available in the Order preview page for all Genexus order configurations).
Known limitations:
While we have worked on fixing all known issues with the White Glove copy amplicon feature, unpredictable exceptions might still apply preventing users to obtain the desired subset design. In case of issues, please open a bioinformatics support ticket (Regional contact links: Americas, EMEA, Greater China, South Asia, Japan), providing the ID of the White Glove panel of origin and the list of amplicons that needs to be copied over. Resolutions requiring new White Glove design submissions will be considered for prioritization over existing projects based on available resources, but prioritization cannot be guaranteed.
The Ion AmpliSeq Designer Help documentation is currently under revision for the latest features and will be updated in future AmpliSeq Designer releases.
7.6.8 (May 23, 2023)
dbSNP database version update
We have updated our support for dbSNP database to version 155 for both hg19 and hg38 human reference genomes.
Support for older dbSNP versions is not available, however users can always enter genomic coordinates if obsolete dbSNP annotations are needed.
Known limitations:
Exceptions might apply to the copy amplicon feature for human White Glove designs preventing users to obtain the desired subset design. While we are working on fixing this. In case of issues with copying White Glove amplicons please open a bioinformatics support ticket (Americas, EMEA, Greater China, South Asia, Japan), providing the ID of the White Glove panel of origin and the list of amplicons that needs to be copied over. Resolutions requiring new White Glove design submissions will be considered for prioritization over existing projects based on available resources, but prioritization cannot be guaranteed.
The Ion AmpliSeq Designer Help documentation is currently under revision for the latest features and will be updated in future AmpliSeq Designer releases.
7.6.7 (March 21, 2023)
Link to Thermo Fisher Scientific Analytical Validation (AV) Consulting Services
In this release we have introduced direct access to the Thermo Fisher Analytical Validation Consulting Services info page and to the form to request to be contacted by an AV Specialist for additional information. Links are available within banners in the login page and in the top bar of all pages once user is logged in, as well as a “Need help with panel validation process?” link within all panel pages.
Thermo Fisher Scientific AV Consulting Services are meant to accelerate and streamline your validation process, including all custom Ion AmpliSeq™ and Ion AmpliSeq™ HD panels, by providing technical project management of your research lab’s AV, potentially reducing your overall time to launch by 75%.
Note: AV Consulting availability and levels of service may vary by region. Please contact an AV Specialist at the provided link for more specific information.
New look & feel of the Help section
We have renewed the look & feel of the Help section for a better navigation experience. Once navigating to the Help section, the Ion AmpliSeq Designer Help, Frequently Added Questions, User Guides, and Release Notes items and submenus are now always accessible in the side bar, while the content of the selected item is dynamically displayed on the rest of the page.
Support of the new Ion AmpliSeq™ HD Library Kit with HD Enhancer
The newly released Ion AmpliSeq™ HD Library Kit with HD Enhancer (Cat. No. A57283), bringing improved performance to the manual library preparation of Ion AmpliSeq HD panels (Gene Studio workflow), is now available in the list of recommended consumables in the Order preview page for custom HD panels. It replaces the Ion AmpliSeq™ HD Library Kit (Cat. No. A37694) as the recommended library preparation kit.
Known limitations:
The new look & feel for the Help section is currently available only when accessed after login. Access from the login page will be implemented in a future release.
The Ion AmpliSeq Designer Help documentation is currently under revision for the latest features and will be updated in future AmpliSeq Designer releases.
7.6.6 (February 1, 2023)
Extension of the copy amplicon functionality to human DNA White Glove panels
From this release, amplicons belonging to DNA human White Glove (WG) designs can be copied to new or existing draft designs to generate full/partial subset Made-To-Order panels with WG content*.
Both the source and the destination design must belong to the same AmpliSeq.com account, and both designs must have the same human reference (hg19 or hg38). WG amplicons can be copied directly from the WG panel page (only design-to-design through the “Copy Amplicons” button). Additional gene, region, amplicon targets can (optionally) be added to the design draft before submission to obtain the new subset panel. Made-To-Order subset panels with any WG content can NOT be shared with other users.
*Exceptions might apply, which will require a new full WG submission to obtain the desired subset design.
Extension of copy amplicon functionality from Ion AmpliSeq™ HD to Ion AmpliSeq™ Made-To-Order designs
Users can now copy amplicons from Ion AmpliSeq HD to Ion AmpliSeq (human, hg19) Made-To-Order designs in addition to the already existing ability for the opposite process. For compatibility reasons, only amplicons within the overlapping size range of AmpliSeq HD and standard AmpliSeq chemistry (i.e., 125 to 175 bp) can be copied in this fashion.
Known limitations:
Copying White Glove amplicons with amplicon ID length >30 characters is not available at this time. This limitation will be fixed in an upcoming release. In case of such an occurrence, please open a bioinformatics support ticket (Regional contact links: Americas , EMEA, Greater China, South Asia, Japan) indicating the White Glove panel of origin and providing a list of amplicons that need to be copied over.
The Help documentation is currently under revision for the latest features and will be updated in future AmpliSeq Designer releases.
7.6.5 (November 24, 2022)
Hotspot target prioritization and 2-pool hotspot solutions
Users can now optionally prioritize targets when creating custom Ion AmpliSeq™ and Ion AmpliSeq™ HD hotspot designs.
When only overlapping amplicons are available for covering close-by targets in a 1-pool hotspot design, the design pipeline will eliminate one or more of them to resolve the conflict, resulting in the missed coverage of the input targets they would have covered.
The new hotspot target prioritization functionality launched in this release (available for hotspot designs only, see FAQ for more details) allows users to define a priority which hotspot target(s) the design pipeline should attempt to retain when such overlapping amplicons conflicts occur. The corresponding amplicons - if available - will preferentially be kept in the final design. This feature aims at maximizing the coverage of the hotspot targets that are the most relevant for the user.
Hotspot target prioritization is not meant for increasing the overall coverage of 1-pool hotspot designs; in case, lower specificity solutions (see FAQ) or 2-pool hotspot solutions (see below) are better suited for the scope.
Starting from this release, 2-pool solutions are also automatically generated for hotspot designs. These solutions allow maximizing coverage over the input targets in presence of overlapping amplicon conflicts and are made available whenever the 1-pool hotspot solutions do not cover all input targets.
The new hotspot target prioritization and 2-pool hotspot solutions features combined provide users with a wider choice of solutions to cover their hotspot targets of interest.
The feature to propose as Community panel is back again. A new "Propose as Community panel" button is now available in the design page of any human DNA or RNA custom Made-To-Order AmpliSeq™ and AmpliSeq™ HD panel. This functionality allows users to propose custom panels to be considered for sharing as AmpliSeq™ Community panels directly from the panel page by filling-in an online form with some basic information.
Only custom panels previously ordered and verified for performance are eligible for being considered as Community panels. Evidence (e.g., publication, datasets, etc.) of the actual panel performance, along with indications on any protocol modifications and/or specific analysis setup applied, will be required.
Proposals are subject to review and approval by Thermo Fisher Scientific and submitters will be contacted with further details on the next steps once their proposals will have been evaluated.
Known limitation:
The Help documentation is currently under revision for the latest features and will be updated in future AmpliSeq Designer releases.
7.6.4 (October 2, 2022)
Search & Compare improvements
The following improvements were introduced for the Search & Compare tool:
- Keyword search capability extended to non-human Community panels
- Reference to design name added to Search Results Table (Panel ID/Design panel link column and panel Details pop-up) and to Panel Comparison page
- Product type info added to Details pop-up (Search Result Table) and to Panel Comparison page
- Wet-lab coverage availability info (yes/no) added to Panel Comparison page
- Minor bug fixes and improvements
New option to include oncology hotspot.bed file in panel result files
We introduced a new feature in the Start a new AmpliSeq design page for human DNA Made-to-Order panels (gene, hotspot and subset) to optionally include an oncology hotspot.bed file in the panel result files.
The hotspot.bed file will contain a curated list of oncology hotspots whose coordinates overlap with the designed amplicons. To learn more about hotspot.bed files and their use in variant analysis, visit the AmpliSeq FAQ.
Note: The underlying curated hotspot list might be subject to updates in future AmpliSeq release versions. In case, notification of any version change will be provided in the corresponding Release Notes.
COSMIC database version update to v94
- We have updated our support for the COSMIC database to version 94 for both hg19 and hg38 human reference genomes.
- Support for older versions of COSMIC will not be available, however users can always enter genomic coordinates if obsolete annotations are needed.
- For future reference, starting from this release, COSMIC version supported in ampliseq.com at the time of design will be reported in the header line of "submitted", "designed" and "missed" output bed files for hotspot panels ("cosmicVersion" tag).
Removed 384-well plate option for AmpliSeq HD RNA fusion panels
Creating sub-panels by picking individual oligos from 384-well plates for AmpliSeq RNA fusion designs is not supported due to fusion assay design properties. Therefore, in alignment with Made-to-Order standard RNA fusion panels, the Tubes Only format is now the only option available when ordering AmpliSeq HD RNA fusion panels.
Known limitation:
The Help documentation is currently under revision for the latest features and will be updated in future AmpliSeq Designer releases.
7.6.3 (August 3, 2022)
Redesigned Fixed Panels dashboard
We have redesigned the Fixed Panels dashboard for enhanced user experience in searching, accessing, and managing Community and Ready-to-Use Panels.
Improvements include:
- Simplified table layout, including only essential information and actions. Additional details remain available on each panel page, which will be accessible by clicking on the panel name link.
- Replaced the “Download panel files” button with an inline download icon ()
- Column sorting and dynamic filtering capability (plus the ability to clear all applied filters at once).
- According to panel type and size, an inline button for Order Panel, Preview Order, or Generate SOS gets displayed.
- Revisited search and filter panel with filter categories now, matching the order of table column headers.
Known limitation:
The Help documentation for the Redesigned My Design and Fixed Panel dashboard is currently under revision and will get updated in a future AmpliSeq Designer release.
7.6.2 (June 29, 2022)
Redesigned My Designs dashboard
We have revised the My Designs dashboard layout to make it clearer and improve user experience with accessing and managing custom designs.
Enhancements include:
- New “Delete Selected” button that enables deleting one or more selected designs at once. Warning! If designs shared with collaborators are deleted, they will also be deleted from all collaborators’ accounts. The delete action cannot be undone.
- Ability to edit panel Name and Details directly from the dashboard table ( icon in the Design column)
- Column sorting and dynamic filtering capability (plus the ability to clear all applied filters at once)
- “DNA”, “RNA” and “DNA and Fusions” sub-tabs replaced by the new filterable “Pipeline” column in dashboard table. All panel types per product line are now listed in the same table for easier access
- New filterable “Status” column added to dashboard table incorporating the design status and the design editable/locked icon. The corresponding “Status” top filter has been removed from the dashboard.
End of support for Internet Explorer
Starting from this version, AmpliSeq.com is not supported on Internet Explorer (any version). Users are encouraged to switch to an alternative supported browser to access AmpliSeq.com.
7.6.1 (May 11, 2022)
Improved clarity on relaxed parameter solutions as a strategy to improve design in silico coverage
When a Made-To-Order design is submitted, the AmpliSeq pipeline computes several alternative solutions aimed at covering the required targets. Once the design is completed, only the High Specificity solution (recommended result) is presented to the user.
Increased coverage over this standard solution might have been achieved in one or more alternative solutions where amplicon design specificity constraints were iteratively relaxed. If such solutions exist, the user can add them to the available default solution clicking on the “Improve coverage” button in the panel page.
Since this approach has demonstrated to be useful to many customers, starting from this version we provide more detailed information on how the amplicon design specificity strategy works to increase in silico coverage over missed targets so that users can make more educate decisions in choosing these options.
In particular, information content now clearly specifies that only amplicons overlapping the regions missed by the standard solutions are subject to specificity relaxation, while the remaining amplicons in the design maintain their default high specificity level. Consequently, although lower specificity may result in increased off-target amplification for these amplicons, the effect is limited to the regions close to those missed by the default solution and not to the whole design.
New “Trouble Signing In” link on AmpliSeq.com login page
A “Trouble Signing In” link on AmpliSeq.com login page points users to a new dedicated AmpliSeq FAQ section where solutions to the most common scenarios preventing users to sign in into their AmpliSeq.com account are provided.
Enhanced handling of Expression Controls in custom RNA fusion panels
Starting from this version, the pre-populated list of 12 gene expression assays in your AmpliSeq Made-to-Order Fusion designs is fixed (the default genes cannot be removed). As in previous versions, the gene list can be expanded to include further gene expression assay content. The 12 default genes are therefore always included in panel result files where they are configured as Expression Control assays for downstream analysis/QC purposes in Ion Reporter.
Note: Starting from v7.6.2 (Expected release date- end of June 2022), AmpliSeq.com will not be supported on Internet Explorer (any version). Please plan to switch to an alternative supported browser to access AmpliSeq.com.
7.6 (February 27, 2022)
A new Search & Compare tool is now available at the top of the Home page. The tool replaces the previous basic search tool and allows users to perform advanced content searching by keyword or by gene list throughout all panels available in their account. Searchable content includes custom DNA solutions, custom gene expression RNA solutions, designs shared by collaborators, Community panels and Ready-To-Use panels whose content is hosted on AmpliSeq.com (e.g. Cancer Hotspot Panel v2, Comprehensive Cancer Panel, etc.).
Search results are provided directly on the user interface as a list of panels matching the search criteria, organized in a sortable and filterable table.
The main details of each matching panel (e.g., formats available, number of pools, number of amplicons, application area, etc.) are available directly in the result table (“Details” button), together with direct links to access the corresponding panel page for a more detailed review and ordering of the panels.
Up to three panels in the search result list can be selected for a side-by-side comparison over their main features.
The “Search by Keyword” option allows searching designs available in the user account based on keywords, gene names, or symbols (free-text), performing an exact string match search (case insensitive) against panel descriptions, panel names, and gene names included in panel target list.

By default, results are provided as a list of the top 10 panels (with the option to show all results) matching the search criteria and ranked by relevance. Results can be filtered/sorted according to different criteria.
The “Search by Genes” option allows searching designs available in the user account based on their gene content. Search terms can be manually entered, pasted, or uploaded (CSV file) as a list of gene names, gene symbols, or synonyms (case insensitive). A mandatory Sample Criteria selection in the tool interface (“cfDNA”, “FFPE” or “Standard” for DNA panels; “Gene Expression” for RNA panels) focuses the search only to the panel's intended use and allows for a streamlined review of search results.

By default, results are provided as a list of the top 10 panels (with the option to show all results), matching the search criteria and ranked by the percentage of matched search genes. A detailed list of matched/missed genes is provided for each hit and results can be filtered/sorted according to different criteria.
The design of new custom panels of the relevant type given the selected sample criteria and based on the searched gene list can be launched directly from the search result page.
When comparing search-by-gene results (up to 3), the main details of each selected panel are provided side-by-side on the user interface, including the total number of genes (exportable as file) and the number of amplicons, the list of matching/missing genes, the number of pools, etc. A list of all matched and missed genes across compared solutions can also be exported as an Excel file for easier review.
RefGene version annotated in panel files
Starting from this release, when a new custom design is created the RefGene version used to perform the panel design will be annotated in the header line of the panel bed files (“refGeneVersion” tag) for future reference.
Known limitation
The Help documentation for the Order Journey process and instrument selection options is currently under revision and will be updated in a future AmpliSeq Designer release.
7.4.12 (December 20, 2021)
Instrument compatibility updated to align to Genexus Software 6.6
The instrument selection options available in the Order Preview journey for all panels have been updated to align with products/solutions officially supported in Genexus Software 6.6.
General guidelines on instrument support by panel type have been updated accordingly also in the “Ion Torrent Genexus System – Consumables” section in the AmpliSeq FAQ documentation.
Description of IGV tool tracks added to AmpliSeq On-Demand panels page
A brief description of the tracks shown in IGV Lite tool for AmpliSeq On-Demand panels is now directly accessible within the panel page by clicking on the information icon located on the top-right corner of the IGV tool.
Known limitation
The Help documentation for the Order Journey process and instrument selection options is currently under revision and will be updated in a future AmpliSeq Designer release.
7.4.11 (November 9, 2021)
Order Preview workflow improvements
In this release, we have updated the Order Preview workflow to allow for an easier order configuration of all AmpliSeq custom panels, while also providing a more informative review of the selected panel format before proceeding with ordering.
The main improvements include:
- Streamlined workflow with less pop-ups and information tailored to each panel type.
- A renewed Order Preview page shown by default at the end of the panel order configuration process. The page provides a detailed summary of the configuration chosen for the panel (exportable in text format as SOS document if needed) before proceeding with the order.
Among other things, the info displayed on the Order Preview is meant to help with:
- planning experiments, e.g., details on the provided materials plus the direct link to relevant User Guide for volume usage per sample
- ordering, e.g., detailed part number and quantity (if needed for offline orders), bundled kits (if any), additional recommended consumables (optional)
Ordering format and special instructions for White Glove panels are displayed in details at the preview order stage within AmpliSeq.com, according to requirements pre-defined at the White Glove design request phase.
Note: The details provided in the Order Preview page are meant to enhance the user experience while configuring custom AmpliSeq panels for ordering. The ordering process, as well as the format, provided materials, manufacturing process and content of all panel types remain otherwise unchanged.
Inquiries related to AmpliSeq ordering can be sent by email to the following addresses (by region):
FAQ search case-insensitive
The FAQ search functionality is now case insensitive to allow for a more effective searching experience.
In-page AmpliSeq On-Demand panel finalization
The Finalize Design functionality for AmpliSeq On-Demand panels is now executed within the panel page, removing the need to go through the panel order configuration journey.
Other minor bug fixes
Known Limitations:
- Availability of Genexus at the instrument selection step in the Order Preview (as well as in FAQ and Help documentation) is based on the compatibility with Genexus Software 6.2.1. Alignment with Genexus Software 6.6 compatibility will be introduced in the next AmpliSeq Designer release.
- The Help documentation for the Order Journey process is currently being revised and will be updated in a future AmpliSeq Designer release.
7.4.10 (July 29, 2021)
RefGene version update for hg19/hg38
Gene annotation employed for target submission by gene name (Made-To-Order designs) is now updated to RefGene v98 for hg19 and v99 for hg38.
Gene annotation of AmpliSeq On-Demand panels remains unchanged at RefGene v74 to maintain consistency with pre-designed content. AmpliSeq On-Demand spike-in designs also remain unchanged at RefGene v74 for compatibility reasons.
Three of the genes included in the Oncomine Tumor Specific inventory (TCEB1, HIST1H1E and HIST1H3B) have updated gene symbols (but otherwise unchanged coordinates) in RefGene v98. These genes are now reported with their updated gene symbols (ELOC, H1-4 and H3C2, respectively) in any new or unlocked designs (locked designs will maintain v74 gene symbols). Old gene symbols are still available as aliases / synonyms.
SOS document improvement
The SOS document content was revisited for addressing bug fixes and introduce usability improvements, including:
- Simplified Account information section.
- Improved Panel information section (pool content format, amplicon size range, panel type description).
- Enhanced clarity for Oncomine Tumor Specific Panels (bundled library kit and panel content description).
- Improved Order information section (added panel part number description, clear indication of quantity to be used in manual orders).
- Fixed incorrect part number reported for some AmpliSeq HD DNA solutions.
- Fixed missing panel part number for panels with more than 15,000 amplicons.
- Fixed missing special instructions for White Glove and Ready-To-Use panels.
- Fixed minimum order quantity for Made-To-Order panels with less than 48 amplicons (96 primers).
- Other minor bug fixes.
AmpliSeq On-Demand Finalize design workflow
The Finalize Design functionality is now available also for AmpliSeq On-Demand panels. Once finalized, designs are locked from further editing and panel files (including BED files) are directly available for download. Only locked designs can be ordered, once configured in the desired format.
Notes: only genes selected when finalizing a design will be included in the final design and editing of locked designs is possible after creating a copy.
Known Limitations:
Embedded special instructions for White Glove designs, if any, are now automatically captured by the SOS document. However, the Preview Order workflow on the user interface does not show nor matches with embedded special instructions (if any) associated with White Glove designs. User interface improvements on special instructions handling for White Glove designs are in progress and will be made available in a future release.
7.4.8.3 (April 5, 2021)
Standard-Order-Sheet (SOS):
- To improve the customer experience with order processing, new Standard Order Sheet (SOS) is added for all the product types. This document will be available for download during the checkout process in AmpliSeq designer at the new SOS pop-up, Order Summary pop-up and Pre-cart page. This document will include the user account details, panel and order related details to facilitate in the order process.


New Help documentation:
- New and updated help documentation have been added to AmpliSeq.com, to help with the various product types (Oncomine tumor specific panel, AmpliSeq On-Demand, Made-to-Order and AmpliSeq HD).
7.4.8 (January 20, 2021)
New pricing availability during the checkout process
Pricing information will now be available during the checkout process in AmpliSeq Designer without the need to proceed to the thermofisher.com Cart. List pricing will be displayed to customers at the Order Summary popup and the Pre-Cart page where additional optional consumables are available for selection.
Clarification of use of FFPE designs with AmpliSeq HD
FFPE designs with amplicon ranges between 125-175bp, also referred as “FFPE long”, may only be used with the 530 chip used by the Ion GeneStudio™ sequencing instruments. Designs with amplicon ranges between 75-125bp, which are considered “short”, are now considered to be “dual use” and may be used with all chips (530, 540, 550 and GX5), which includes the new Ion Torrent™ Genexus™ Integrated Sequencer. Dual use refers to the sample input compatibility which can be DNA extracted from liquid biopsy samples or FFPE stored material.
Support for the LDB3 gene for the GRCh38 genome reference
The gene LDB3 is now added to our GRCh38 genome reference. This can now be accessed by creating a new draft and typing the key word “LDB3” as the gene symbol.
- Ion AmpliSeq™ MH-74 Plex Research Panel
- Ion AmpliSeq™ VISAGE (Visible Attributes through Genomics)-Basic Tool Research Panel
- Ion AmpliSeq™ PhenoTrivium Panel
7.4.7 (December 28, 2020)
Inclusion of fusion detection from RNA for select Oncomine™ tumor specific panels.
All solid Oncomine tumor specific panels for cancer research focused on solid tumors can now be selected with a Solid Tumor RNA panel that enable detection of fusions with FusionSync. The Solid Tumor RNA panel cannot be selected with the Oncomine Lymphoma Research panel.
Ability to create custom Oncomine™ tumor specific DNA panels from gene list.
Easily create a new custom Oncomine tumor specific DNA panel from a gene list. Genes that are available in inventory, will be included in a new custom panel. All others will be listed in a downloadable file as either not in stock or invalid if the gene symbol is not recognized. The user will be able to either type the genes directly or upload them using a CSV file.
Updated FAQs available for the Oncomine tumor specific panels.
New and updated FAQs have been added to the Oncomine tumor specific panel help section, to help understand the behavior of this new functionality.
Improvements in in-silico coverage for Spike-in panels created for AmpliSeq On-Demand designs.
As part of our continuous improvement efforts, we have optimized the in-silico coverage for Spike-in panels created for AmpliSeq On-Demand designs.
7.4.6.4 (November 19, 2020)
Data base upgrades
Versions for the dbSNP are being updated for the hg19 and mm10 reference genomes. For hg19 the dbSNP version is now v153 and for mm10 the dbSNP version is v150.
New Research Area categorization for the AmpliSeq Fixed panels
To simplify the organization of all our panels with fixed content (Community and Ready-to-Use panels), we have reduced the number to the following 5 categories and preserving the DNA and RNA top level categories:
- Cancer Research Panels
- Genetic and Complex Disorders Research Panels
- Infectious Disease Research Panels
- Microbial Research Panels
- Other Research Panels
7.4.6.3 (November 10, 2020)
New Oncomine tumor specific panel for HRR research
A new predesigned panel called Ion Torrent™ Oncomine™ Homologous Recombination Repair Pathway Predesigned Panel has been added to our collection of Oncomine tumor specific panels.
7.4.6 (September 24, 2020)
Official support for genomic references downloaded from the NCBI website.
To improve the ease of working with Custom References, AmpliSeq Designer has adopted the NCBI genomic reference format, which allows any reference downloaded from the NCBI site to be uploaded to AmpliSeq.com without the need to further edit the file. In addition, contig names are now officially supported up to 200 characters in length, which covers most names found in references available from the NCBI.
Flexibility for addition or removal of an existing polymorphism BED file associated to a Custom Reference.
In previous versions, users wanting to add or edit the polymorphism BED file had to re-upload the associated custom reference and provide the new BED files at the time of upload. With this release, we now enable the addition or editing of the polymorphism BED file, without the need to re-upload the custom reference.
New FAQs available to explain transfer and sharing rules for designs associated to Custom References.
A new section called "Ion AmpliSeq Designer – Custom References" has been added to our FAQs. This section describes the expected behavior when a design that is associated to a Custom References is transferred or shared to another user.
Official support of the SARS-CoV-2 panel for the Ion Torrent Genexus System.
The Ion AmpliSeq SARS-CoV-2 Research Panel is now officially supported for the Ion Torrent Genexus System. The instrument selection option to include the Genexus System has been added as well for the Preview Order process.
"Instant Survey" available for users to optionally answer brief questions from the AmpliSeq Designer team.
To help with gathering feedback from our users, we have implemented a new survey tool where we will be asking a few brief questions. The survey is strictly optional, and participation is greatly appreciated.
7.4.5 (July 7, 2020)
Improvements in in-silico coverage for AmpliSeq HD designs and single solutions per design now available.
Our newest design improvements for in-silico coverage, are now available for custom AmpliSeq HD designs. The magnitude of the improvement will vary from design to design and is dependent on the amplicon size and number of pools.
In addition, following the same practice as with custom AmpliSeq designs, the user will be prompted to select the “DNA Type” based on 2 amplicon sized options. Option 1 is for DNA with amplicon size range of 75-125bp, which has been optimized for cfDNA input material. Option 2 is for amplicon size range of 125-175bp, which works best for DNA sourced from FFPE material.
Enhanced design logic to reduce primer-dimer interactions for AmpliSeq HD designs.
Advancements in our design logic have been incorporated to help reduce the likelihood of primer-dimer for designs with 500 amplicons per pool or less. Suspected amplicons likely to cause primer-dimer will be automatically shifted from one pool to the other (in the case of 2 pool designs) or removed completely from the design. In the case where amplicons have been copied from a design using the “Copy Amplicons” function, a file named “filtered.tsv” will be created to provide in-sight into the detected conflicting must have amplicons. The user will have the option to resolve the conflict by selecting one of the listed amplicons and re-running the creation of their design.
New AmpliSeq On-Demand genes available.
Additional 18 genes have been added to our inventory. Designs containing these genes will be automatically updated if they’re in the Results Ready state. For designs that have been ordered or locked, there will be a note to the user providing them with the guidance to clone the design in order to access the newly inventoried content.
New FAQs available to better understand the logic around Copy Amplicons.
Specific FAQs have been added to explain the behavior when copying amplicons from a specific DNA Type (e.g. FFPE) to an existing draft design with a pre-defined DNA type (e.g. cfDNA). Visit the “Ion AmpliSeq Designer How To”, for more information regarding rules around the Copy Amplicons function.
7.4.3 (April 28, 2020)
Improved ordering journeys for panels and option to select consumables.
We have improved our ordering process to better reflect the options that the user has when ordering panels. Depending on the selected instrument, the library preparation method and the ordering format (e.g. tubes or tubes plus 384-well plates), the user will be provided with a new summary and the option to browse additional consumables. This improvement should make it easier to select the necessary consumables during the check out process.
Known Limitations:
- The chip calculator is not working properly for the Genexus System. From the main header it is not listing the estimated libraries per chip. From the design, it is not being selected properly.
- The approximation for number of reactions for Spike-In panels is only listing the lower limit. This number depends on the target parent panel. A fix to clarify the number of reactions depending on the parent size is needed.
7.4.2 (March 4, 2020)
Improvements in in-silico coverage for designs using Custom References and Extended Genomes and single solutions per design now available
We're extending our newest design improvements in in-silico coverage to designs being created from custom references or any of our pre-loaded genomes (e.g. Cow, Sheep, etc.) The magnitude of the increase will vary from design to design, and heavily depends on amplicon size and the number of pools.
Following further user feedback, we have incorporated a new parameter that allows the user to create design solutions for only the application of interest. This is captured as the "DNA Type" and provides the user with 4 amplicon sized options. Option 1, is for DNA with amplicon size range of 125-140bp. Option 2, is for designs with amplicon range of 125-175bp. Options 3 and 4, are for high quality DNA and gives the user the option for 125-275bp or 125-375bp amplicon range solutions respectively.
Improved designs and turn around time for commonly designed genes
For our commonly designed genes, we have pre-loaded improved design solutions that combine automated and semi-automated methods to quickly generate designs with the highest possible quality and in-silico coverage.
Addition of tumor type associations to the Oncomine tumor specific panels
For our Oncomine tumor specific panels, each gene in the panel now contains tumor type associations to help with interpretation.
Design sharing improvements:
We have now enabled bulk sharing of a design to multiple users, with an automated email notification alerting the recipients that a design has been shared to their account.
Login process and Access Code:
The login process has changed. Users need to provide their login credentials in a separate page prior to accessing the Home Page. This change is done as part of our continuous improvement of our internal systems to enhance the security of our site, and compatibility with single-sign-on.
In addition, a new mechanism for generating an Access Code has been provided under the "My Account" options. This Access Code will now be used for importing panel information in Torrent Suite and Ion Reporter.
Known Limitation:
Sharing of designs originating from a Custom Reference, is currently not supported. This feature will be supported in an upcoming release.
7.4 (January 15, 2020)
Introducing the new Oncomine™ tumor specific panels for Cancer Research:
- Content: 10 new tumor specific panels leveraging Ion AmpliSeq™ technology.
- Recommended system: Ion AmpliSeq 530 chip with 8 samples per chip for DNA only.
- Analysis: Dedicated Ion Reporter workflows.
- Reporting: Supported by Oncomine Reporter.
- Library compatibility: Compatible for use with Ion AmpliSeq™ Library Kit Plus and Ion AmpliSeq™ Library for Chef DL8.
- Flexibility: Ability to customize the panel desing by removing genes or adding genes from inventory. Panels can be created with up to 150 genes.
New Design Sharing management system:
A new system for managing the sharing of designs has been created. The new management tool makes it easier to keep track which designs have been shared, and to whom they've been shared to. The recipient of the design must be registered in ampliseq.com for their email address to be recognized, and only one email can be input at a time. Support for bulk sharing to multiple emails will be provided in a future release.
Login process:
The login process will be changing. Users will provide their login credentials in a separate page prior to accessing the Home Page. This change will be done as part of our continuous improvement of our internal systems to enhance the security of our site, and as a pre-requisite for future single-sign-on compatibility with corporate systems where available.
7.2 (June 26, 2019)
New Home Page to welcome our existing and new users.
- A new Home Page has been created to help our users navigate our products. The new Home Page organizes the products as "In stock" or "Made-to-order" to help set the proper expectations on product delivery time. The page also includes the most commonly inquired product features in table format, so users can quickly find the solution that best fits their needs.
Improvements in in-silico coverage now available.
- As part of our continued efforts for improving our automated design creation algorithms, we have made improvements that on average achieve an overall increase in in-silico coverage for our designs. The magnitude of the increase will vary from design to design, and heavily depends on amplicon size and the number of pools. Our internal testing indicates that this can range from ~1% to about ~41% in in-silico coverage improvement. On rare cases, we observed a small decrease in coverage, however this was for around 1% of the total designs tested. Correlation between the in-silico design and the in-vitro performance remain the same. The mentioned improvements are only available for AmpliSeq Made-to-Order (Custom) and for genomes hg19, hg38 and mm10. Improvements to AmpliSeq HD and custom references based designs, are expected later in the year.
Focus on creation of single solutions based on application area.
- Per user feedback, we have incorporated a new parameter that allows the user to only create design solutions for only the application of interest. This is captured as the "DNA Type" and provides the user with 4 options. Option 1, is for cell-free DNA with amplicon size range of 125-140bp. Option 2, is for the FFPE optimized designs with amplicon range of 125-175bp. Options 3 and 4, are for high quality DNA and gives the user the option for 125-275bp or 125-375bp amplicon range solutions respectively. As part of this effort, users should observe an overall decrease in design time, given that only the relevant solutions for the application of interest will now be created.
Increase in the Ion AmpliSeq HD multiplex limit.
- The Ion AmpliSeq HD technology's new multiplex limit is now set at 5,000 amplicons per pool. This represents a 10X increase in multiplexing capability from last year, which should translate into greater design flexibility for our users. Designs containing more than 500 amplicons per pool up to the new limit of 5,000 amplicons per pool, will now be orderable from our website.
Known Issues:
- The number of exons in the coverage summary does not add up correctly in the case of specific overlapping genes (e.g. DAZ2, DAZ3, DAZ4).
7.1 (May 27, 2019)
Additional AmpliSeq On-Demand genes are now available:
- New wet-lab tested genes have been added to our inventory selection.
- Existing designs will be automatically updated to reflect the newly available genes.
- Ordered designs can be cloned to create a design containing newly available genes.
Updated Disease Research Area associations for AmpliSeq On-Demand Genes based on new genes added to inventory.
7.0.7 (March 17, 2019)
Issue with Ion AmpliSeq HD 384-well plate files is now fixed
An issue with our 384-well plate files for all designs created for the Ion AmpliSeq™ HD technology was identified and is now fixed. This issue affected all designs with the prefix “IAH”, and was responsible for the incorrect mapping of the oligo location within each well. The 384-well plate files for designs created prior to March 17, 2019, will be fixed on a rolling basis. If you have ordered an Ion AmpliSeq HD design in the 384-well plate format before March 17, and you wish to do partial pooling, please contact your local support team or reach out to our global support for further guidance on how to deal with this issue.
Introducing the new Ion AmpliSeq™ Research Panels:
7.0.6 (December 16, 2018)
New alert for AmpliSeq On-Demand designs containing greater than 275bp amplicons
- A small alert will now be displayed for designs containing greater than 275bp amplicons.
- From the alert the user can determine if additional flows are required for complete end-to-end coverage of the longest amplicon in their panel.
New histogram for visualizing the amplicon size distribution for all designs
- A new histogram will be available from the user interface for users to visualize the amplicon distribution content.
- The image will be available for download, as well as part of the download zip bundle.
COSMIC database version update to v86
- We have updated our support for the COSMIC database to version 86.
- Support for older versions of COSMIC will not be available, however users can always enter genomic coordinates if obsolete annotations are needed.
Increased limit for simultaneous design requests
- As of version 7.0.5 we have increased the limit from 2 to 5 simultaneous designs created.
- A higher limit enables the flexibility to our users to get to their ideal design faster.
7.0.5 (October 30, 2018)
Improved design range for cfDNA solutions for AmpliSeq HD:
- The amplicon range for cfDNA solutions generated for AmpliSeq HD designs has been further optimized to increase in-silico coverage.
- The new range is now from 75-125bp amplicons for the AmpliSeq HD cfDNA solutions.
- Older cfDNA solutions (75-100bp) will no longer be created, but solutions that were previously created will remain accessible in the user's account.
New "Special Instructions" option for increased manufacturing flexibility for AmpliSeq Made-to-Order designs. Instructions include:
- Ability to synthesize at 5X.
- Synthesis at 50X for Spike-in use (BED file modifications still need to be performed by the user. Consult your local FBS for help).
- Larger scale option for pooled tubes only at either 2X or 5X concentration.
- Option for plates only orders (no pooled tubes to be included).
7.0.4 (September 14, 2018)
Additional AmpliSeq On-Demand genes are now available:
- New wet-lab tested genes have been added to our inventory selection
- Existing designs will be automatically updated to reflect the new content
- Ordered designs have the option to be cloned to access the newly added content
7.0 (June 16, 2018)
This release of Ion AmpliSeq Designer unveils a brand new product called “Ion AmpliSeq™ HD technology” and includes new capabilities geared towards achieving ultra-high sensitivity and customization:
New Ion AmpliSeq HD Made-to-Order panels offer:
- Ion AmpliSeq HD panels leverage molecular tags to achieve ultra-high levels of sensitivity.
- Ion AmpliSeq HD panels are only compatible for use with the new Ion AmpliSeq HD Library Kit (SKU: A37694) and Ion AmpliSeq HD Dual Barcode Kit 1-24 (SKU: A37695).
- The Ion AmpliSeq HD panels are fully customizable and are only supported for the hg19 genome at this time.
- Input for these panels is the same as traditional Ion AmpliSeq Made-to-Order panels. Input can be provided using:
- Gene symbols (names)
- Gene regions as defined by genomic coordinates (Start and End)
- SNP ID’s using COSMIC or dbSNP definitions
- A new design type for cfDNA is now offered with an exclusive amplicon range of 75-100bp. The new amplicon range reflects design optimization that has been done for cfDNA using the Ion AmpliSeq HD chemistry. The previous range 125-140bp for cfDNA will not be offered for Ion AmpliSeq HD; however, it will remain available for regular Ion AmpliSeq designs.
New set of Ready-To-Use Panels:
- Oncomine™ Tumor Mutation Load Assay
- Oncomine™ Myeloid Research Assay
- Oncomine™ Focus Assay
- Oncomine™ Comprehensive Assay v3
- Oncomine™ BRCA Research Assay
- Oncomine™ TCR Beta-SR Assay
- Oncomine™ TCR Beta-LR Assay
- Oncomine™ Immune Response Research Assay
New User Interface layout:
- Our site is now organized based on the chemistry that supports the multiple applications.
- On the left side the Ion AmpliSeq technology is hosted, and the different applications are organized in tabs.
- On the right side the new Ion AmpliSeq HD technology is found, and the button to start a new Ion AmpliSeq HD is provided.
- Additional enhancements such as mouse hover over on the top bar allow easy navigation to the desired designs.
- The designs page is organized in 3 categories: 1) On Demand, 2) Made-to-Order and 3) Ion AmpliSeq HD. The new organization provides a better way of finding your designs by product category or by underlining chemistry.
- Made-to-Order and Ion AmpliSeq HD categories also offer a quick button to display DNA or RNA designs only.
Known Issues:
- Chip Calculator: Do not display “Approximate # Library Samples/Chip” table for Ion Gene Studio Instrument and 550 chip for S5/S5 XL Instrument. Please find below table as reference for the same.
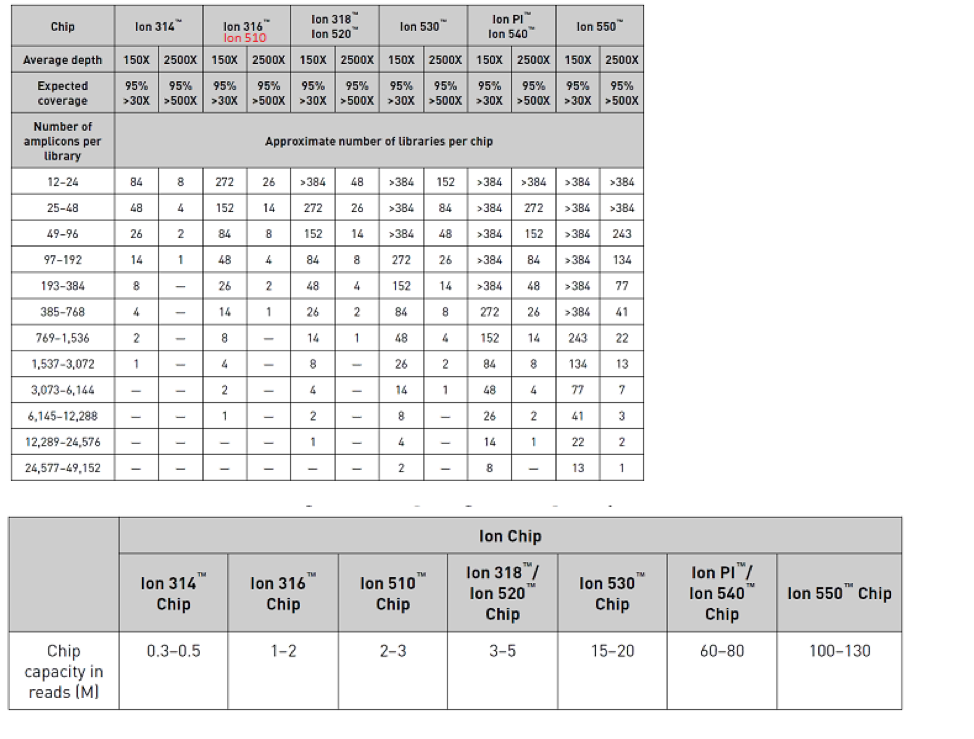
- Copy amplicons from Fixed Panels into AmpliSeqHD designs is not allowed in this version of AmpliSeq Designer. Adding Amplicons from Fixed Panel using Amplicon ID would result into error on UI.
6.1.4 (January 15, 2018)
Support for our new Ion GeneStudio S5, S5 Plus and S5 Prime Sequencers and 550 chip.
- The new Ion GeneStudio Sequencers and 550 chip were launched on Jan-08-2018, and with this update we’re providing full compatibility within Ion AmpliSeq Designer. The new Ion GeneStudio Sequencers and 550 chip represent a new exciting flexible offer for high performance bench-top NGS solutions.
Updates to the plan.json file for the Ion AmpliSeq Exome Panel to support data from 540/550 chip.
- New updated parameters for the Ion AmpliSeq Exome Panel are now available through a new version of the plan.json file, for improved variant calling on data from 540/550 chip.
6.1.3 (January 03, 2018)
Additional new AmpliSeq On-Demand genes are now available:
- Newly added wet-lab validated genes have been incorporated to our selection
- Existing designs will be automatically updated to reflect the new content
- Ordered designs have the option to be cloned to access the newly added content
6.1.2 (December 5, 2017)
Introducing The New Ion AmpliSeq Research Panels
Ion AmpliSeq Transcriptome Mouse Gene Expression Research Panel
- The Ion AmpliSeq Transcriptome Mouse Gene Expression Research Panel is used to measure expression levels of 23,930 RefSeq genes using only 1 pool. The panel targets 20,767 well annotated RefSeq genes and 3,163 coding and non-coding genes with provisional annotation based on Genome Reference Consortium Mouse Reference 38 (GRCm38). All amplicons are of approximately 150 bases in length. We recommend sequencing 8 samples on a Ion 540 Chip or Ion PI Chip.
Ion AmpliSeq Circular RNA Research Panel
- Circular RNAs (circRNAs) were recently discovered as a novel class of noncoding RNA that are widely expressed in various tissues. Although the function of the vast majority of circRNAs remains unknown, several of them have already been associated with human disease. Currently, there is lack of essential approaches to study circRNAs. A target-enrichment sequencing method suitable for high-throughput screening of circRNAs and their linear counterparts in large sample sets were developed using AmpliSeq technology.
6.1 (October 15, 2017)
New AmpliSeq On-Demands genes are now available
- Additional wet-lab validated genes are now accessible
- Existing designs will be update to reflect new content
- Ordered designs can be cloned to benefit from updated content
New larger size packs now available
- 96 reactions for Manual workflow
- 32 reactions for Ion Chef
Ordering of larger AmpliSeq On-Demand designs is now supported
- Designs with up to 500 genes (or 15,000 amplicons, whichever occurs first) can now be ordered
- Affordable pricing offered for large designs containing 301-500 genes (check the Cart to learn more)
- A new tool has been created to quickly search inventory status of genes without the need to create a design
6.0 (May 22, 2017)
This release of Ion AmpliSeq Designer unveils a brand new product called “Ion AmpliSeq On-Demand” and includes an impressive array of new features:
New Ion AmpliSeq On-Demand panels offer:
- The benefit of accessing verified content with the flexibility of customization. The new Ion AmpliSeq On-Demand panels have been optimized and verified similarly to our Ready-to-Use panels, while offering gene level customization for flexible panel creation.
- Visualization of wet-lab verified data is now readily accessible through our new IGV Lite tool. Click on a gene to access the expected coverage data at the amplicon level without having to use an external tool like the UCSC genome browser.
- A new visualization mode known as the “Grid view” has been created to help you review the design of your Ion AmpliSeq On-Demand panel, and to facilitate adding or removing content.
- Once you’re ready to order, order your panel in small reaction packs offered at affordable prices. The small reaction configuration of the Ion AmpliSeq On-Demand panels helps lower the price per sample, making custom gene panels more accessible than ever.
- With the Ion AmpliSeq On-Demand panels, a new tool has been developed to allow you to browse our catalog of genes that have been organized based on their association to various disease research categories.
- A multi-level browsing experience enables users to select genes starting with their specific research areas of interest.
- Selection of Disease Research Area categories can be made at any level, just keep in mind that up to 800 genes can be selected per panel for design creation and of that, 300 genes can be ordered per panel at this time.
New User Interface layout:
- Our site is now organized based on content rather than by design workflow.
- You can now easily browse the Ready-to-Use panels, select genes for On-Demand panels, or access the Made-to-Order panel workflow which enables our traditional custom design capabilities for ultimate flexibility.
- The menu bar on top of the page has been simplified allowing you to hover over the quick access buttons to see your designs in the Draft, Results Ready or Ordered stages.
- A new Notifications tool is also available to provide you the latest important communications from the Ion AmpliSeq Designer team.
Known issues:
After an On-Demand design is created, genes added using the Add Gene tool are not categorized correctly when using the “Group by: Disease Research Area” function. Currently the genes will be categorized under “Added” rather than their respective Disease Research Area group.
During the design review stage, the “Group by: Disease Research Area” option should list all the genes that have been pre-selected under the original Disease Research Area category. In the future, this will include both the genes that are available and not available in our On-Demand catalog. However, only the genes that are available in our On-Demand catalog will appear under this category at this time, while the unavailable genes will appear in the “Unclassified” category.
At this time, when an On-Demand panel is cloned, the genes that are available On-Demand will be re-selected while genes that are not available will be deselected by default. Preserving the user selections and de-selections after using the cloning option is currently not supported.
The filtering functionality in the IGV tool which allows users to view specific genomic positions based on Expected Coverage values is not working as expected at this time. Data ranges do not get correctly filtered.
When an incorrect gene symbol is uploaded via the Gene List option in the homepage or via the Upload file option after a panel has been already created, the user will have the option to download the list of unrecognized genes as a file. The downloaded file should contain the “.CSV” extension; however our current download does not include this extension. The user will need to add the CSV extension to the file and correct the gene symbol, prior to being able to upload the list into the Ion AmpliSeq Designer website.
5.6 (August 1, 2016)
New support for creating custom designs using the GRCh38 (hg38) genome.
- We now support creation of custom designs using the latest publicly available human genome called hg38. Please note that support is limited to the consensus contig, and a new alternate contig containing the GSTT1 gene. This gene will no longer be located in Chr22 but on the new alternate contig. All other alternate contigs are not supported in this release.
Updates to the plan.json file for the Ion AmpliSeq Exome Hi-Q panel.
- New updated parameters for the Ion AmpliSeq Exome Hi-Q are now available through a new version of the plan.json file, for improved variant calling.
- The new Ion AmpliSeq BRCA Reflex, Hereditary Cancer Research Panel is a 2 pool panel with 610 amplicons, and contains 25 hereditary cancer-related genes which have a central role in DNA repair and mismatch repair pathway.
- The new Ion AmpliSeq BRCA Plus, Extended Hereditary Breast and Ovarian Research Panel, is a 3 pool panel with 699 amplicons that targets 11 genes known to harbor mutations related to breast and/or ovarian cancer.
New set of RNA Pathway panels has been added to our collection of pre-designed RNA research panels
- Ion AmpliSeq RNA MAPK Pathway Panel - this panel is involved in Cell Cycle & Proliferation, Virology and Signal Transduction.
- Ion AmpliSeq RNA Human Cancer Pathways Panel - this panels includes a large number of genes that are involved in DNA repair, angiogenesis, cell adhesion and extracellular matrix, as well as cell cycle and apoptosis.
- Ion AmpliSeq RNA Inflammation Response Panel - this panels targets numerous genes that are taking part in key pathways of inflammation response that include chemokines.
- Ion AmpliSeq RNA WNT Signal Pathway Panel - this panel includes a list of genes that are responsible for the regulating the production of WNT signaling molecules.
- Ion AmpliSeq RNA Pancreatic Adenocarcinoma Panel - this is a panel that targets genes known to be over-expressed in Pancreatic carcinoma.
- Ion AmpliSeq RNA Breast Cancer Panel - this is a very comprehensive panel targeting over 1,100 genes involved in breast cancer development and progression.
5.4.2 (June 27, 2016)
New automated in-silico histogram for custom designs.
- As part of our output files, we now include an in-silico histogram publication quality image to help visualize the amplicon insert size distribution of our custom designs. The image includes on the X-axis the Amplicon Insert Size in bp units, and the Amplicon Count on the Y-axis. Information on the total number of amplicons in the design and their size range is also provided.
5.4.1 (May 18, 2016)
Additional new Ion AmpliSeq™ Research Panels for Inherited Diseases Research Area.
- Ion AmpliSeq Cardiac Arrhythmias and Cardiomyopathy Research Panel
- Ion AmpliSeq Pulmonary Research Panel v2
- Ion AmpliSeq Dysmorphia-Dysplasia Research Panel v2
- Ion AmpliSeq Inborn Errors of Metabolism Research Panel v2
- Ion AmpliSeq Primary Immune Deficiency Research Panel v2
- Ion AmpliSeq Endocrine Research Panel v2
- Ion AmpliSeq Gastrointestinal Research Panel v2
- Ion AmpliSeq Deafness Research Panel v2
- Ion AmpliSeq Renal Research Panel v2
- Ion AmpliSeq Inherited Cancer Research Panel
- Ion AmpliSeq Epilepsy Research Panel
- Ion AmpliSeq Autism Research Panel
- Ion AmpliSeq Ovarian Cancer Research Panel
- Ion AmpliSeq Dermatology Research Panel v2
These new panels provide greater focus for research and expand the breath for which the Ion AmpliSeq technology can be used for. To order, please use the “Add to Cart” or “Request quote” or “Request Information” buttons. For the request information button, you will need to provide your contact details and a representative of our team will be in touch shortly.
5.4 (April 20, 2016)
- Internal bug fixes and administrator enhancements
5.0 (October 6, 2015)
Improved workflow based user experience
- Our new Ion AmpliSeq Designer 5.0 has been improved to bring a workflow based approach to creating, reviewing and ordering your designs. With this new version, you will find that navigating through the application has never been easier, and getting to where you need to go is only a click away.
Access to Ion AmpliSeq Research Panels with no log in
- Our Ion AmpliSeq Research Panels known as Ready-to-use and Community Panels, have been reorganized by default to fit within common research areas where targeted sequencing is used.
- You will now find direct links from the home page to important research areas which you may review without the need to register or log in. Once you’re ready to order, you will need to sign in to your account and proceed with the ordering process.
Support for our new Ion S5™ and S5™ XL Sequencers
- As a reminder, the new S5 and S5 XL have been launched on September 1, and we provide full compatibility within Ion AmpliSeq Designer. The new S5 and S5 XL systems represent a new exciting offer for high performance bench top sequencers.
New trial sizes available for 4 Inherited Diseases Research panels and the Pharmacogenomics Research panel (Due early to mid-October)
Update: Due to inventory backlog, the availability of trial sizes for the following research panels will be for early to mid-October:
- Ion AmpliSeq™ Cardio Panel – To be available in 8 reaction and 32 reaction Chef-ready-trial kits.
- Ion AmpliSeq™ Hematology Panel – To be available in 8 reaction and 32 reaction Chef-ready-trial kits.
- Ion AmpliSeq™ Neurological Panel – To be available in 8 reaction and 32 reaction Chef-ready-trial kits.
- Ion AmpliSeq™ Ophthalmic Panel – To be available in 8 reaction and 32 reaction Chef-ready-trial kits.
- Ion AmpliSeq™ Pharmacogenomics Research Panel – To be available in only 32 reaction Chef-ready.
Special pricing for Custom Gene Fusion panels
- Based on customer demand, for a limited time, our Custom Gene Fusion panels will continue to be priced the same as traditional RNA Gene panel designs.
- As a reminder, the variant analysis workflow for Custom Gene Fusions is exclusively provided by Ion Reporter™. For more information please visit the Ion Reporter™ site here.
4.4.10 (September 15, 2015)
Support for our new Ion S5 and S5XL Sequencers
- Full compatibility of AmpliSeq Designs
- S5/S5XL family of chips fully integrated
- A new exciting offer for high performance bench top sequencers
New trial sizes available for 4 Inherited Diseases Research panels and the Pharmacogenomics Research panel.
The following recently released panels are now available in trial sizes for easier customer access:
- Ion AmpliSeq™ Cardio Panel – Available in 8 reaction and 32 reaction Chef-ready-trial kits.
- Ion AmpliSeq™ Hematology Panel – Available in 8 reaction and 32 reaction Chef-ready-trial kits.
- Ion AmpliSeq™ Neurological Panel – Available in 8 reaction and 32 reaction Chef-ready-trial kits.
- Ion AmpliSeq™ Vision Panel – Available in 8 reaction and 32 reaction Chef-ready-trial kits.
- Ion AmpliSeq™ Pharmacogenomics Research Panel – Available in only 32 reaction Chef-ready
Introducing the new Ion AmpliSeq™ Research Panels
Ion AmpliSeq™ Ebola Research Panel
- The new Ion AmpliSeq Ebola Panel was developed as a quick, accurate and cost effective method to identify the present of Ebola virus (EBOV). The panel contains 145 amplicons for targeting the Ebola gene.
Ion AmpliSeq™ TB Research Panel
- The new Ion AmpliSeq™ TB Research Panel used for targeting genes associated with antimicrobial resistance is now available. This panel consists of 109 amplicons in 2 pools to provide a quick, accurate and cost effective means to identify variants in 8 genes (embB, eis, gyrA, inhA, katG, pncA, rpoB, rpsL) associated with antimicrobial resistance in Mycobacterium tuberculosis (TB).
Ion AmpliSeq™ Pharmacogenomics Panel
- The new Ion AmpliSeq™ Pharmacogenomics Panel is a single pool panel of primers used to perform multiplex PCR for the preparation of amplicon libraries from genomic “hot spot” regions that play a critical role in DME genes. By focusing on 136 well documented SNP and indel variants that captures CYP2D6 copy number variations at both the gene level and the exon 9 rearrangement, the panel enables screening of a broad selection of haplotypes including *36. This targeted, ready-to use panel provides high value pharmacogenomics content while remaining flexible by allowing user customization.
Ion AmpliSeq™ Inhered Diseases Research Panels
- We have added a collection of 13 new panels involved in various Inherited Disease areas to complement our existing offerings in this field. This new menu of Ion AmpliSeq Research Panels offers a broad selection of targeted panels that enable researchers to survey across collective sets of genes that have been scientifically curated as the most likely suspects for containing a candidate mutation based on criteria such as phenotype and disease. Panels represent a wide range of inherited conditions from cardiovascular to dermatological disease. Use of our Ion AmpliSeq™ Technology enables coverage of up to 766 genes employing a straightforward and rapid PCR based workflow requiring as little as 10 ng of input DNA. These panels are comprehensively designed and can be used as-is or can be easily customized with the power of Ion AmpliSeq™ Designer to create phenotype based focused sub panels. This collection of panels are 2-pool and have over 99% coverage.
Browse Panels without logging in
- The Ready-to-use and Community Panel interface has been redesigned to allow visitors easier access to our family of pre-designed panels. Site visitors can review the content of our panels without the need to register or log in to an Ion AmpliSeq™ Designer account.
Special pricing for Custom Gene Fusion panels
- For a limited time, new Custom Gene Fusion panels will continue to be priced the same as traditional RNA Gene panel designs.
Analysis workflow exclusively provided by Ion Reporter™
- Variant analysis workflows for Custom RNA Gene Fusion designs are exclusively provided by Ion Reporter™. For more information please click visit the Ion Reporter™ help section here.
4.4.9 (September 1, 2015)
Support for our new Ion S5™ and S5™ XL Sequencers
- Full compatibility of AmpliSeq Designs
- S5/S5 XL family of chips fully integrated
- A new exciting offer for high performance bench top sequencers
Introducing the new Ion AmpliSeq™ Research Panels
Ion AmpliSeq™ TB Research Panel
- The new Ion AmpliSeq™ TB Research Panel used for targeting genes associated with antimicrobial resistance is now available. This panel consists of 109 amplicons in 2 pools to provide a quick, accurate and cost effective means to identify variants in 8 genes (embB, eis, gyrA, inhA, katG, pncA, rpoB, rpsL) associated with antimicrobial resistance in Mycobacterium tuberculosis (TB).
Ion AmpliSeq™ Pharmacogenomics Panel
- The new Ion AmpliSeq™ Pharmacogenomics Panel is a single pool panel of primers used to perform multiplex PCR for the preparation of amplicon libraries from genomic “hot spot” regions that play a critical role in DME genes. By focusing on 136 well documented SNP and indel variants that captures CYP2D6 copy number variations at both the gene level and the exon 9 rearrangement, the panel enables screening of a broad selection of haplotypes including *36. This targeted, ready-to use panel provides high value pharmacogenomics content while remaining flexible by allowing user customization.
Ion AmpliSeq™ Inherited Diseases Research Panels
- We have added a collection of 13 new panels involved in various Inherited Disease areas to complement our existing offerings in this field. This new menu of Ion AmpliSeq Research Panels offers a broad selection of targeted panels that enable researchers to survey across collective sets of genes that have been scientifically curated as the most likely suspects for containing a candidate mutation based on criteria such as phenotype and disease. Panels represent a wide range of inherited conditions from cardiovascular to dermatological disease. Use of our Ion AmpliSeq™ Technology enables coverage of up to 766 genes employing a straightforward and rapid PCR based workflow requiring as little as 10 ng of input DNA. The panels can be used as-is or can be further customized through Ion AmpliSeq™ Designer to easily remove or add additional amplicons.
Browse Panels without logging in
- The Ready-to-use and Community Panel interface has been redesigned to allow visitors easier access to our family of pre-designed panels. Site visitors can review the content of our panels without the need to register or log in to an Ion AmpliSeq™ Designer account.
Special pricing for Custom Gene Fusion panels
- For a limited time, new Custom Gene Fusion panels will continue to be priced the same as traditional RNA Gene panel designs.
Analysis workflow exclusively provided by Ion Reporter™
- Variant analysis workflows for Custom RNA Gene Fusion designs are exclusively provided by Ion Reporter™. For more information please visit the Ion Reporter™ site here.
4.4 (May 28, 2015)
Support for RNA Gene Fusion designs
- RNA Fusion designs creation has never been easier. Simply input and select your fusion(s) of interest and let Ion AmpliSeq™ Designer do the rest.
- Support for gene fusion detection through highly targeted AmpliSeq™ assays is done using an extensive library of known fusion transcripts.
Special pricing for Custom Gene Fusion panels
- For a limited time, new Custom Gene Fusion panels will be priced the same as traditional RNA Gene panel designs.
Panel search user interface revamp
- The Ready-to-use and Community Panel search user interface has been redesigned to enhance the user experience and allow easier discovery of panels of interest.
Analysis workflow exclusively provided by Ion Reporter™
- Variant analysis workflows for Custom RNA Gene Fusion designs are exclusively provided by Ion Reporter™. For more information please click visit the Ion Reporter™ help section here.
Known Limitations
- Import of custom gene fusion panel files into Torrent Suite™ 4.4.3 will fail. Users are encouraged to create a generic run plan in Torrent Suite™ for any custom gene fusion run.
- Full support of custom gene fusions will be available in Ion Torrent Suite™ 4.6. Please stay tuned for release updates.
4.2.3 (February 16, 2015)
Support for cell free DNA designs
- Create designs for cell free DNA (cfDNA) targets with our new 140bp amplicon solutions.
- Ion AmpliSeq™ RNA Lung Fusion Cancer Research Panel
- Ion AmpliSeq™ CFTR Panel
- Ion AmpliSeq™ TP53 Panel
- Ion AmpliSeq™ Colon and Lung Cancer Research Panel v2
- Ion AmpliSeq™ Hearing Loss Research Panel v1
4.2.2 (January 30, 2015)
Customize your panels!
- Now the Ion AmpliSeq™ AML Research Panel and the Ion AmpliSeq™ Comprehensive Cancer Panel join the list of our panels that can be customized.
Upload even larger design reference files!
- Our Custom Reference up-loader can support 2 GB uploads. However, users are encouraged to contact us to upload files of up to 4.0 GB if needed.
4.2 (December 18, 2014)
Customize your panels!
- Re-use amplicons of interest from AmpliSeq Community Or Ready-to-use panels
- Re-use amplicons of interest from previous AmpliSeq panels
- Add new targets and mix new amplicons and re-used amplicons to form a new panel
New pre-loaded genomes now available!
- Chinese Hamster (criGri1)
- Dog (canFam3)
Upload larger design reference files!
- Users may now upload files of up to 2.0 GB.
3.6 (July 11, 2014)
New Custom Design Functionality!
- New variant ID support for Hotspot designs
- Add variants to your design using dbSNP or COSMIC IDs
- Designer will automatically convert variant IDs to hg19 or mm10 coordinates
- Provides option to enter either coordinates or supported variant IDs
New Ready-to-Use Panels!
- A25642 - HID-Ion AmpliSeq™ Ancestry Panel (released June 27, 2014)
- Number of amplicons: 168
- Number of pools: 1
- Input DNA: 1ng per tube
- A25643 - HID-Ion AmpliSeq™ Identity Panel (released June 27, 2014)
- Number of amplicons: 124
- Number of pools: 1
- Input DNA: 1ng per tube
- Ion AmpliSeq™ RNA Apoptosis Panel (updated June 23, 2014)
- Panel size: 267 genes
- Number of amplicons: 267
- Number of pools: 1
- Input RNA: 10ng
- Updated BED file
- Ion AmpliSeq Hearing Loss Research Panel v1 (released June 30, 2014)
- Panel size: 294.81kb
- Coverage: 95.7378913
- Number of amplicons: 2064
- Number of pools: 2
- Input DNA: 20ng
- Ion AmpliSeq™ Colon and Lung Research Panel v2 (released May 28, 2014)
- Panel size: 14.5kb
- Coverage: 100
- Number of amplicons: 92
- Number of pools: 1
- Input DNA: 10ng
3.4 (March 31, 2014)
- Ion AmpliSeq™ TP53 Panel
- Covers all coding exons (no UTRs)
- 24 amplicons, 2 pools, 125-175bp design
- Compatible with FFPE samples
- Ion AmpliSeq™ CFTR Panel
- Covers all protein coding regions including UTRs, intron/exon boundaries (20bp), 162 CFTR2.org mutations including 23 ACMG/ACOG mutations, four deep intronic mutations, and two large deletions (CFTRdele2,3 and CFTRdele22,23).
- 102 amplicons, 2 pools, 125-225bp design
New Custom Design Functionality!
- New hotspot design capability
- Improved design algorithm to optimize coverage with a single-pool solution
- Previous tiling methodology would favor multiple pool solutions to cover nearby hotspots. With current methodology, the algorithm looks for an optimal single pool solution avoiding amplicon overlap.
- Hotspots are optimally positioned on amplicon insert for more robust variant calling
- The new algorithm seeks to position the hotspot close to the center of the amplicon. The benefit is that reads are mapped with higher reliability, and a greater probability of having reads from the forward and reverse strands, fulfilling the requirement for variant calling.
- Updated RefSeq
- More up-to-date human RefSeq is used for defining gene targets
- The human RefSeq has been updated to the latest reference available in January, 2014 (RefSeq v63). This provides an updated reference for gene design.
3.2 (February 7, 2014)
New Look and Feel!
- Improved dashboard
- Quick access to your custom designs, Ion Panels and Community Panels
- Notifications on the progress of your custom designs
- Quick access to support resources
- Powerful search/filter capabilities to quickly find pre-designed panels.
The HID-Ion AmpliSeq™ Identity Panel is one of the first Next-Generation Sequencing (NGS) solutions for human identification that provides discrimination of individuals similar to STR genotype match probabilities used by forensic analysts.
3.0 (November 4, 2013)
New AmpliSeq™ custom design pipeline
- Get close to 100% coverage on human exome targets
- Rerun your designs to see the improvements
Improve in silico coverage by selecting designs with relaxed primer specificity.
- Choose the recommended design for best on-target % of sequence reads.
- Select the “increase coverage” button and immediately see higher coverage designs, if available.
- These pre-computed designs deliver increased coverage in GC-rich regions, while seeking to minimize off-target amplification.
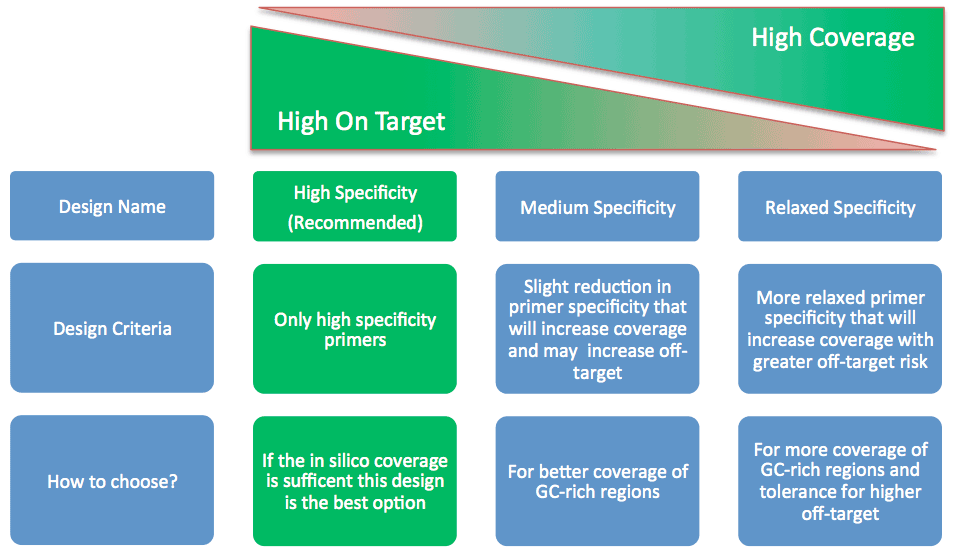
Before: (91.57% coverage)
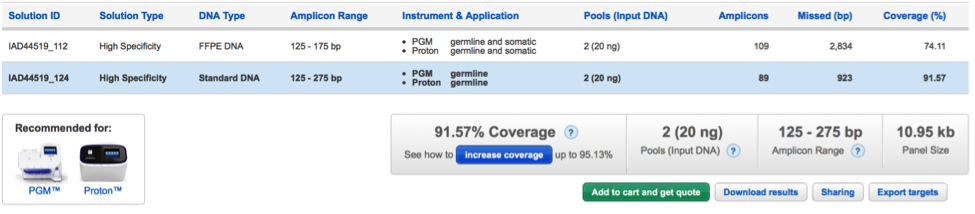
After: (95.13% coverage)
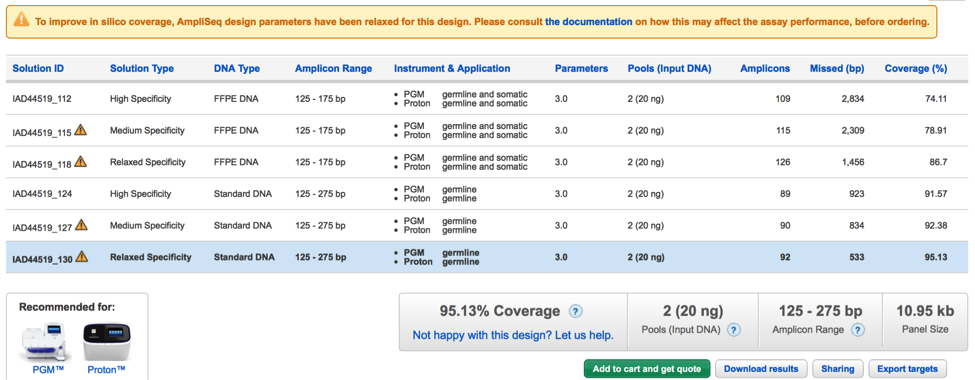
Greatly improved design times
- Smaller, gene-centric designs returned within minutes
- Designs up to 1 Mb within a week
Share your own AmpliSeq custom panels with the world
- See the first such panel, the Dementia Research Gene Panel, under Community Panels.
- Share your own designs using the “Sharing” button:
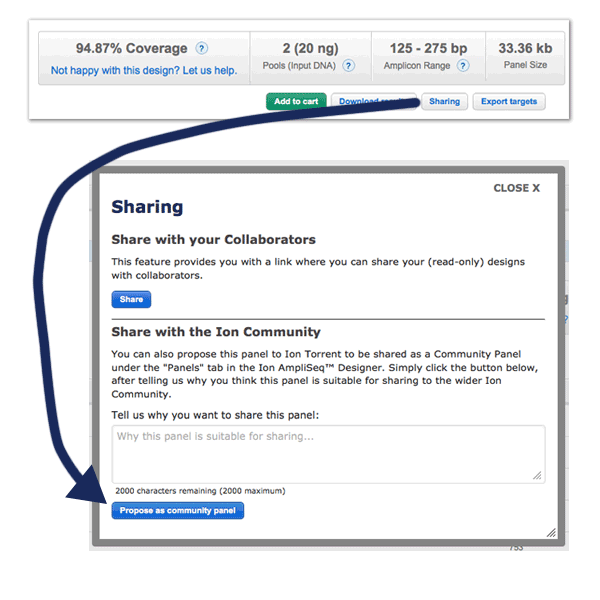
2.2.2 (October 11, 2013)
- Transition to single sign on system on LifeTechnologies.com and improved ordering experience
To streamline the process of ordering Ion AmpliSeq™ Panels, AmpliSeq Designer is transitioning user accounts from Ion Community accounts to Life Technologies accounts.
Get all the details here: Migration Help Page
- Benefits of Transitioning to Life Technologies accounts
In one single checkout process add all the reagent kits and chips needed to complete your AmpliSeq experiment
Order all AmpliSeq panels using the e-commerce solution on Life Technologies website
See the details of the new experience: Ordering Process
2.2.1 (August 9, 2013)
RNA Panels
2.2 (June 27, 2013)
DNA Panels
Introducing the Ion AmpliSeq™ Exome Panel panel.
This is available under the "DNA Ready-to-use" panel tab.
- Comprehensive coverage of the protein-coding exons

Increased in silico coverage for all standard DNA designs. For better in silico coverage of your existing panels
you will have to redesign them.
- The amplicon range for standard DNA designs has increased to 275bp.
- The same 200bp sequencing kits can be used to sequence the longer amplicons.
Ion Proton instrument supports short designs (125bp-175bp) for germline applications.
2.0.3 (May 22, 2013)
- RNA AmpliSeq Designs now allow up to 1200 amplicons per pool
- DNA design results display specific reasons why a design cannot be confidently covered by an amplicon.
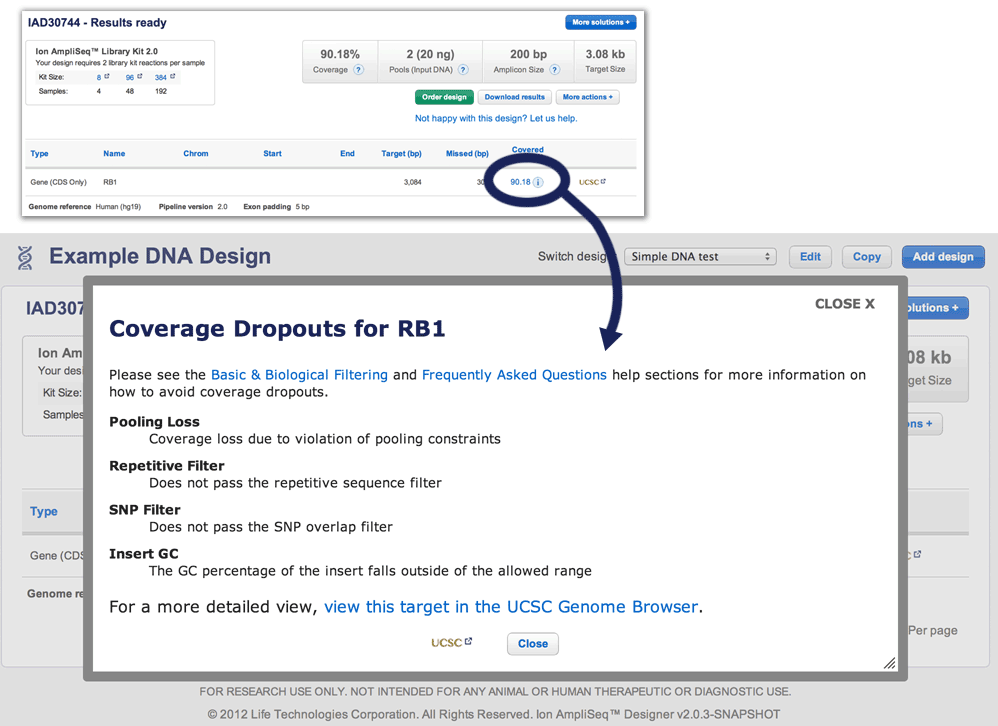
Detailed information can be viewed using the UCSC Browser button.
- Search across ready-to-use and community panels, including the panel name, description, and gene names.

2.0.1 (April 1, 2013)
Ion AmpliSeq™ RNA Ready-to-Use Panels
Two Ion AmpliSeq™ RNA Ready-to-Use Panels have been added to the Panels tab:
- Ion AmpliSeq™ RNA Apoptosis Panel
- Ion AmpliSeq™ RNA Cancer Panel
2.0 (February 17, 2012)
New DNA AmpliSeq™ Custom Pipeline
Custom Mouse Panels
- Target your mouse genes of interest
- The same proven Human design pipeline
Custom RNA AmpliSeq™ Panels
Ready-to-Use RNA AmpliSeq™ Panels
- Ion AmpliSeq™ Colon and Lung Cancer Panel
- Ion AmpliSeq™ BRCA1/2 Panel
Ion AmpliSeq™ Sample ID Panel
A cost-effective and easy-to-use human SNP genotyping panel comprising nine specially designed primer pairs that can be added prior to template amplification to generate a unique ID for each research sample during post-sequencing
analysis.
Ion Library Equalizer™ Kit
Use the Ion Library Equalizer™ Kit to normalize your Ion DNA libraries without the need for dilution or quantification, or the need for any additional equipment.
1.2.9 (December 19, 2012)
You can now order designs that your colleagues have shared with you. Simply use the URL they’ve shared with you to review the design and then click the order button.
Any ordered custom design can now be submitted to Life Technologies for consideration as a Community Panel. To submit one of your designs please complete the form found in the Sharing option under More Actions button on the review page.
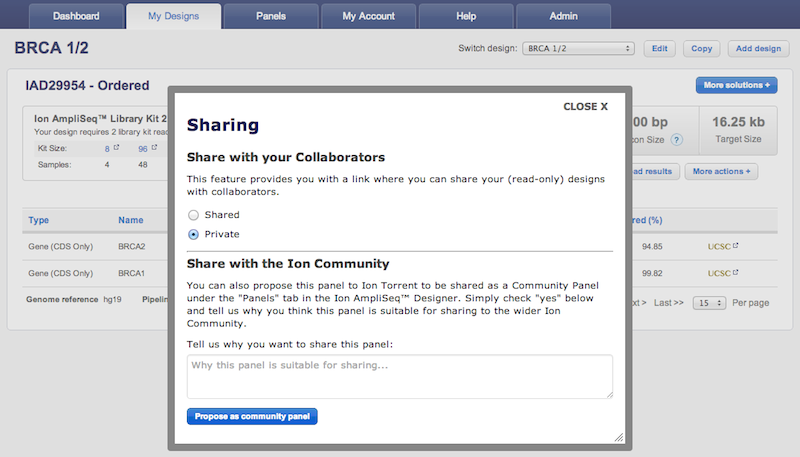
If you haven’t noticed yet, the AmpliSeq™ Training Videos have been updated to include even more great information. Check them and other great resources out under the Help tab. If you’re new to AmpliSeq™ Designer be sure to check out the getting started guide.
After receiving results from the AmpliSeq™ Designer pipeline you can now provide feedback to help us improve the application. You’ll find the feedback link in the top right corner of the review design page.
We’ve also included some minor fixes and user experience improvements.
1.2.8 (November 6, 2012)
- Minor bug fixes and user experience improvements.
- The Ion Panels tab is now called Panels and includes new
AmpliSeq™ Community Panels designed with leading researchers.
AmpliSeq™ Community Panels are currently being testing and verified for performance
and will soon be available for order.
1.2.7 (October 7, 2012)
- Minor bug fixes and user experience improvements.
- Significant performance enhancements to improve turn around time designing your custom assays.
- Profile tab is now called My Account, which has a new Order History page where see all your ordered designs in one place.
1.2.6 (September 10, 2012)
- Custom AmpliSeq™ panels are only compatible with Ion PGM™ 200 Sequencing Kit.
- Introduced a variety of performance enhancements and caching strategies to improve turnaround time.
- Added significantly more help documentation.
Instantly compare designs with both 150bp and 200bp amplicon sizes. Simply click on “More solutions” to compare!
Review, order or customize Ion Ready-to-use Panels.
Share your designs with collaborators.
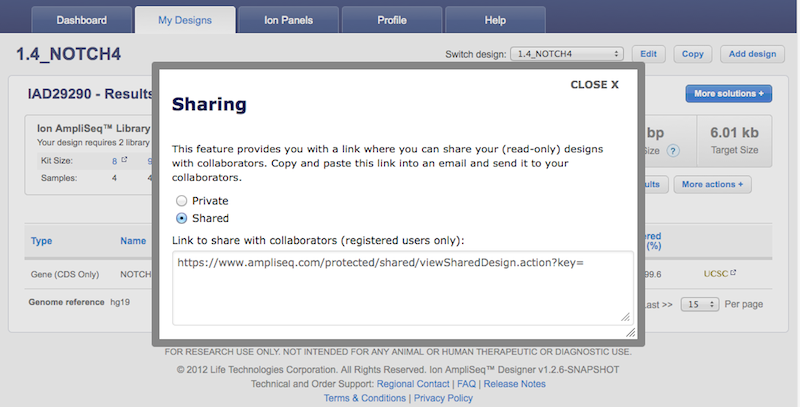
1.2.5 (August 17, 2012)
- Minor bug fixes and user experience improvements.
- Performance enhancements.
1.2.3 (July 27, 2012)
- New designs that have 12-23 minimum amplicons per pool may now be ordered. Existing designs that have 12-23 minimum amplicons per pool must be resubmitted before they can be ordered.
- The icon for the UCSC browser has been updated.
1.2.2 (June 22, 2012)
Many small bug fixes and user experience improvements
1.2 (May 2012)
New features
- Improvements in algorithms and chemistry enable typical exon designs in just 2 pools
- Targeted regions panel designs typically in just 1 pool
- The maximum target size of submission has increased to 1 MB
- Up to 3072 amplicons in a single pool
Integration with UCSC Genome Browser (click on magnifying glass icon in results ready view)

New Dashboard tab to quickly view all your panels, design status and key metrics
Notes








 ), otherwise they are flagged as CNV incompatible (
), otherwise they are flagged as CNV incompatible ( ).
).