Add a custom reference
To create a custom reference, upload a nucleotide sequences FASTA file and an optional polymorphic regions BED file. The FASTA format file contains the context sequence. If there are known polymorphic regions in the FASTA reference sequence, such as SNPs and INDELs, you can upload an optional BED file that contains regions of high variation over which to avoid placing primers for designs against the custom reference. For more information about how the pipeline avoids variant regions during the design process, see Basic and biological filtering.
- In the navigation bar, click Genomes.
- In the Design References screen, click Add reference.
-
In the
Add a custom reference
screen, complete the required information.
Field
Description
Reference name[1]
Enter a unique name for the sequence to be used as a reference for your design. The name can be 3–32 characters in length. The allowed characters are US‑ASCII letters, numbers, spaces, asterisks (*), dashes (-), and underscores (_).
Associated organism for primer specificity check1
Select an associated organism for primer specificity check to improve primer specificity to your custom reference. Primary specificity is improved by favoring primers with few optimal binding sites in the consensus sequence.
Primer specificity check refers to the process of identifying potential primer mispriming events. Primers with high number of potential mispriming events are avoided in panel designs. If you select an associated organism, designs for the custom reference will perform primer specificity check against the preloaded genome sequence of the selected organism. If you select None, primer specificity check will be performed against the custom reference.
The name of the database or source of the reference nucleotide sequence.
Add any notes regarding the custom reference sequence. The description can be up to 2,000 characters in length.
Reference sequences (FASTA) file1
Upload one or more reference sequences FASTA files, or enter FASTA data in a text area. For more information on FASTA format requirements, see FASTA format requirements.
-
To upload a file, click Select file, select the FASTA file of interest from your local storage, then click Open.
Note: The maximum upload file size is 2.0 GB. The limit can be extended up to 4.0 GB on request.
-
To enter or copy/paste FASTA data, click Enter FASTA data in a text area instead, then enter the reference sequence data in the provided field.
Reference short name for Torrent Server1
Enter a name for your reference to be used by Torrent Suite™ Software. The name should be 1–30 characters in length. The allowed characters are lowercase US-ASCII letters, numbers, and underscores (_). For more information on importing custom references into Torrent Suite™ Software, see the Torrent Suite™ Software Help for your version of the software.
A BED format file that indicates regions of the sequences in the custom reference FASTA file with high polymorphism (for example, SNPs, INDELs, or other variations). Ion AmpliSeq™ Designer minimizes primer overlap with these regions. This file is optional. For specifications on creating and formatting BED files for uploading, see Known polymorphism BED file requirements.
To upload a file, click Select file, select the BED file from your local storage, then click Open.
1 Required field -
- When the file upload is complete, click Save.
To confirm the addition of a new custom reference, click Genomes in the navigation bar, then in the Design References screen, click the Custom References tab. For more information, see View available reference genomes.
After a new custom reference is added, you can perform the following actions.
|
Action |
Description |
|---|---|
|
View reference details |
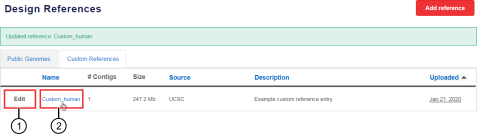
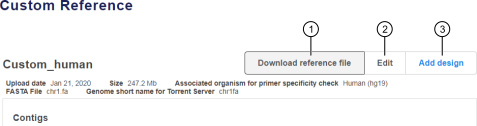
In the Custom References tab, in the Name column, click the link in the row of a custom reference of interest. See callout 2 in Figure 1. The Custom Reference details screen opens, displaying custom reference details, including a summary of contigs and known polymorphisms, if present. See Figure 2. |
|
Edit a reference |
|
|
Download the reference file |
The custom reference ZIP file (Custom_Reference_<reference name>.zip) is automatically downloaded to a designated location in your local storage. |
|
Start a new design using the reference |
The Edit your design screen opens. As you proceed through the design creation workflow, the custom reference is automatically selected as the genome for the design. |