Start a new Ion AmpliSeq™ HD DNA Hotspot design
Ion AmpliSeq™ HD DNA Hotspot designs can include small hotspots (SNP, mutation, and short INDELs). The DNA Hotspot pipeline is optimized to produce single-pool designs for hotspots ≤125 bp.
- In the home screen, in the Ion AmpliSeq™ HD Custom panels pane, click Create Custom HD panel.
- In the Start a new Ion AmpliSeq™ HD design screen, in the Name and details step, enter a design name and optional details.
- In the Application Type step, click DNA Hotspot designs (single‑pool).
-
In the
Select genome to use
step, accept the default
Human (hg19)
selection.
Note: Human hg19 genome reference is the only reference sequence that is allowed with this panel type.
-
Click
Next: Add Targets.
A draft design screen opens (see Figure 1).
-
Add hotspot targets using the options
available in the target addition pane (see callout 1 in Figure 1).
For guidelines on how to add hotspot targets, see Ion AmpliSeq HD Custom panels: DNA hotspot design input specifications.
Option
Description
Add targets manually using SNP ID.
-
In the Add Hotspot tab, in the Type column, select Hotspot by ID.
-
In the SNP ID field, enter a SNP ID. Alternatively, start typing the SNP ID, then select the desired ID from the dropdown list.
-
Click Add target after each entry.
If a target was added successfully, the Target saved successfully message appears. If a SNP ID is invalid, the Target could not be saved error message appears. If needed, you can correct erroneous targets in the targets table, as described in step 7.
Add targets manually using hotspot coordinates.
-
In the Add Hotspot tab, in the Type column, select Hotspot by Coordinates.
-
In the Chrom field, enter the chromosome (for example, enter chr1 for chromosome 1).
-
In the Start and End fields, enter the start and end chromosomal coordinates respectively.
Note: Hotspot size cannot exceed 125 bp.
-
Name (Optional) field, enter an optional name for the region.
Region name cannot contain any of the following symbols: [,;='"\\|/] or spaces.
-
Click Add target after each entry.
If a target was added successfully, the Target saved successfully message appears. If an entry is invalid, the Target could not be saved error message appears. If needed, you can correct erroneous targets in the targets table, as described in step 7.
Upload genomic coordinates of several targets at one time using a SNP list or BED file.
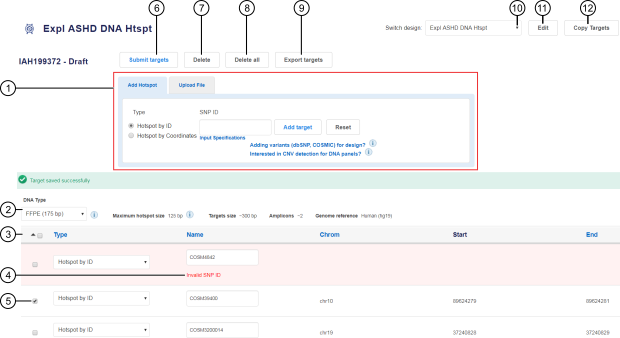 Figure 1. Draft design
screen
Figure 1. Draft design
screen
-
A target addition pane with multiple methods for adding hotspots to a design. Each tab contains fields that are specific to the corresponding target addition method.
-
Select the DNA type.
-
Targets table lists all added hotspots in a design.
-
Targets that did not pass verification are marked in red in the targets table. Fields that contain errors are marked with Invalid and can be corrected.
-
Select the checkbox in the row of each target that you want to delete.
-
Click to submit a design.
-
Click to delete the selected targets (see callout 5).
-
Click to delete all targets.
-
Click to export all targets to a CSV file.
-
Select from the dropdown list to display a different design.
-
Click to edit design name and details.
-
Click to copy targets to a new custom design.
The software uploads the targets, checks them, and verifies regions. The targets are added to the targets table (see callout 3 in Figure 1).
-
-
Review and modify the design as
needed.
Action
Description
Correct erroneous targets
Targets that contain errors are highlighted in red in the targets table. In the row of each erroneous target, modify the fields that are marked as Invalid (see callout 4 in Figure 1). Invalid fields can include Name, Chrom, Start, and End.
Select the DNA type
From the DNA Type dropdown list, select FFPE (175 bp) or cfDNA (125 bp) (see callout 2 in Figure 1.
Note: This setting affects amplicon length in the design.
Delete targets
In the targets table, select the checkbox in the row of one or more targets, then click Delete. To delete all targets, click Delete all. See callouts 5, 7, and 8 in Figure 1.
Change hotspot type
In the targets table, in the row of a target, in the Type column, select a hotspot type from the dropdown list (see callout 3 in Figure 1).
Export targets to a CSV file
Click Export targets. A CSV file that contains the list of all added hotspot targets is automatically downloaded to your local storage (see callout 9 in Figure 1).
You can open the file in Microsoft™ WordPad or Microsoft™ Excel™.
Edit design name and details
Click Edit, then in the Edit your design screen, modify design name and details as needed. Click Save. See callout 11 in Figure 1.
Copy targets to a new design
Click Copy Targets, then in the Copy Design dialog, select the panel type and DNA type, and enter the name and details for the new design. Click Save. See callout 12 in Figure 1.
The new design is added to the list of your custom designs. To view or edit the new design, click in the navigation bar. Alternatively, open a new design by selecting a design from the Switch design dropdown list (see callout 10 in Figure 1). For designs that have been submitted, or locked, the design opens to the Results ready screen.
-
To submit your panel, click
Submit targets,
then click
OK to
confirm your submission (callout 6 in Figure 1.
Up to 5 designs can be submitted at one time.
After submission, a submitted design screen opens, displaying the design confirmation message.
-
Click
OK to
confirm your submission.
When confirmed, you see a confirmation message and receive an email confirming the design submission.
After you submit your design, the targets are locked ( ) and can no longer be edited.
When the assay design results are ready, you receive an e‑mail notification
instructing you to review the results in Ion AmpliSeq™ Designer. Click the
View results
link provided in the email to be directed to the results page. Alternatively, sign
in to Ion AmpliSeq™ Designer (AmpliSeq.com), then in the navigation bar
click to access the results for your design (see My Designs).
) and can no longer be edited.
When the assay design results are ready, you receive an e‑mail notification
instructing you to review the results in Ion AmpliSeq™ Designer. Click the
View results
link provided in the email to be directed to the results page. Alternatively, sign
in to Ion AmpliSeq™ Designer (AmpliSeq.com), then in the navigation bar
click to access the results for your design (see My Designs).
To review design details and place an order, proceed to Order an Ion AmpliSeq HD custom panel.
