Start from a list of dbSNP target identifiers
Use the following example of creating a BED file from dbSNP targets with the UCSC Table Browser (https://genome.ucsc.edu/cgi-bin/hgTables) to generate BED files from other variant formats, such as ClinVar.
-
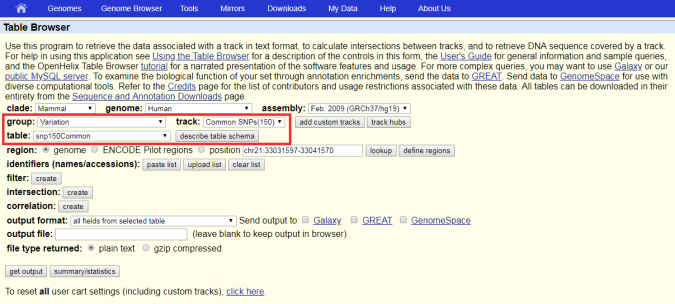
In the UCSC Table
Browser (https://genome.ucsc.edu/cgi-bin/hgTables), make the
following selections from the dropdown lists.
The track and table sections are populated with available SNP databases from dbSNP.
-
In
identifiers (names/accessions)
section, click either
paste list
or
upload list.
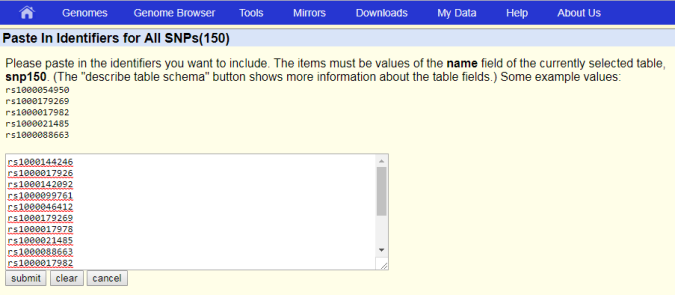
Depending on your selection, Paste in Identifiers or Upload Identifiers screen opens.
- Either upload a text file listing your dbSNP identifiers by clicking Choose File, or paste the list into the provided field, then click submit.
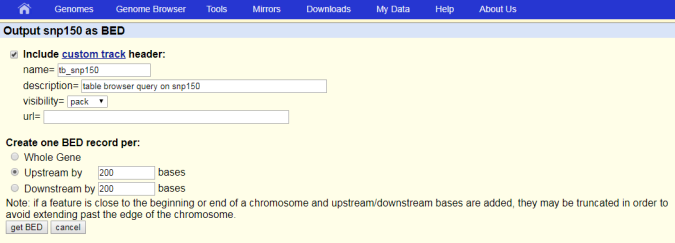
- Click get output.
- In the Create one BED record per: section, select padding with Whole Gene, Upstream, or Downstream options. If needed, adjust the padding value.
-
Click
get BED.
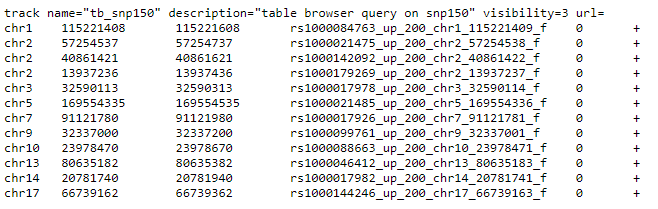
The BED file has the following format, or similar.
×