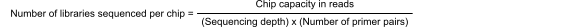Ion Chip capacities for Ion AmpliSeq™ RNA libraries
If there is a user guide for your RNA panel, follow the recommendations in the user guide for your panel.
The following guidelines can be used for panels without a user guide. These recommendations serve as suggestions and the actual capacity to multiplex libraries is determined by the expression levels of the genes included in your Ion AmpliSeq™ RNA panel. The expression levels of the individual genes can vary by input RNA type. We suggest using the formula for new panels and determining actual multiplexing limits empirically.
-
We recommend that you plan for an average of 5,000 reads per amplicon for an Ion AmpliSeq™ RNA library targeting 1–200 genes. The actual sequencing depth that is required depends on the expression levels of the gene targets in your sample RNA, so scale the sequencing depth to accommodate your sample type and research needs.
-
For panels containing fusion detection primer pairs, higher library multiplexing is possible because most targets are not present, and therefore do not create library molecules. For most fusion detection assays, only ~250,000 reads per library are required.
-
Use the following formula and chip capacity table to provide initial guidance for multiplexing RNA‑derived gene expression libraries on Ion sequencing chips.
|
Ion Chip |
|||||||
|---|---|---|---|---|---|---|---|
|
Ion 314™ Chip[1] |
|||||||
|
Chip capacity in reads (M) |
0.3−0.5 |
1−2 |
2–3 |
3−5 |
15−20 |
60−80 |
100–130 |
Chip capacity of Ion 540™ Chip = ~60,000,000 reads
Sequencing depth desired = 5,000 reads per amplicon
Number of primer pairs = 100
60,000,000 / (5,000 × 100) = 120 libraries per Ion 540™ Chip